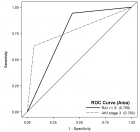
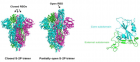
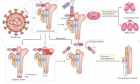
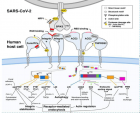
Figure 4
The SARS CoV-2 spike domain, RGD and integrin binding effect-relationship for vaccine design strategy
Luisetto M*, Tarro G, Khaled Edbey, Almukthar N, Cabianca L, Mashori GR, Yesvi AR and Latyschev OY
Published: 20 July, 2021 | Volume 5 - Issue 1 | Pages: 027-041
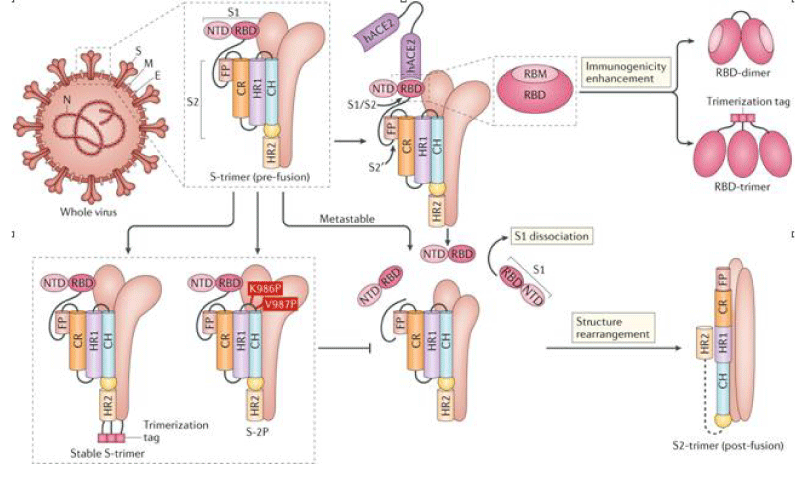
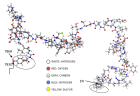
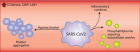
Figure 4:
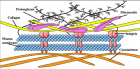
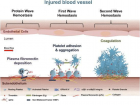
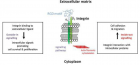
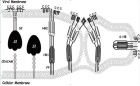
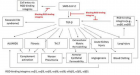
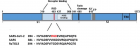
SARS-CoV-2)contains 4 major structure proteins: spike (S), membrane (M) and envelope (E) proteins, which are embedded on the virion -surface, and nucleocapsid (N) protein, which binds viral -RNA inside the virion. The S- protein trimer in its pre-fusion conformation is shown. The S- protein comprises the S1 subunit (which includes the N-terminal domain (NTD) and the receptor-binding domain (RBD)) (the receptor-binding motif (RBM) within the RBD is also labelled) and the S2 subunit (which includes fusion peptide (FP), connecting- region (CR), heptad repeat 1 (HR1), heptad repeat (HR2) and central- helix (CH)). The SARS-CoV-2 S protein binds to its host- receptor, the dimeric human angiotensin-converting enzyme 2 (hACE2), via the RBD and dissociates the S1- subunits. Cleavage at both S1–S2 and S2′ sites allows structural re-arrangement of the S2 subunit required for virus–host membrane fusion. The S2-trimer in its post-fusion arrangement is shown. The RBD is an attractive vaccine- target. The generation of an RBD-dimer or RBD-trimer has been shown to enhance the immune-genicity of RBD-based vaccines. A stabilized S-trimer shown with a C-terminal trimer-tag is a vaccine target. The pre-fusion S protein is generally metastable during in vitro preparations and prone to transform into its post-fusion conformation. Mutation of 2 residues (K986 and V987) to proline stabilizes S- protein (S-2P) and prevents the pre-fusion to post-fusion structural- change.
Read Full Article HTML DOI: 10.29328/journal.apb.1001014 Cite this Article Read Full Article PDF
More Images
Similar Articles
-
The SARS CoV-2 spike domain, RGD and integrin binding effect-relationship for vaccine design strategyLuisetto M*,Tarro G,Khaled Edbey,Almukthar N,Cabianca L,Mashori GR,Yesvi AR,Latyschev OY. The SARS CoV-2 spike domain, RGD and integrin binding effect-relationship for vaccine design strategy. . 2021 doi: 10.29328/journal.apb.1001014; 5: 027-041
Recently Viewed
-
Role of Perianesthesia Nurses in Enhanced Recovery After Surgery (ERAS) Protocols: A Narrative Review and Comparative Outcomes AnalysisOghogho Linda Akarogbe*,Geneva Igwama,Olachi Lovina Emenyonu,Idowu M Ariyibi. Role of Perianesthesia Nurses in Enhanced Recovery After Surgery (ERAS) Protocols: A Narrative Review and Comparative Outcomes Analysis. Int J Clin Anesth Res. 2025: doi: 10.29328/journal.ijcar.1001034; 9: 037-039
-
Climate Change and the Untold Story of EcoanxietyManar Zaki*. Climate Change and the Untold Story of Ecoanxiety. Insights Depress Anxiety. 2025: doi: 10.29328/journal.ida.1001044; 9: 012-016
-
Fiesta vs. Stress Condition the Incidence and the Age at Menarche. Forty Years of ResearchCarlos Y Valenzuela*. Fiesta vs. Stress Condition the Incidence and the Age at Menarche. Forty Years of Research. Clin J Obstet Gynecol. 2025: doi: 10.29328/journal.cjog.1001190; 8: 069-073
-
Minimising Carbon Footprint in Anaesthesia PracticeNisha Gandhi and Abinav Sarvesh SPS*. Minimising Carbon Footprint in Anaesthesia Practice. Int J Clin Anesth Res. 2024: doi: 10.29328/journal.ijcar.1001025; 8: 005-007
-
Identification, Molecular Confirmation, and Antibiotic Sensitivity of Bacteria Isolated from Repeat Breeder Cows in Rangpur DivisionMst. Mousumi Afroj,Md Faruk Islam,Muhammad Mamunur Roshid,Md Shanto Hossain,Md Samiul Tousif,Omiaya Azam Oishi,Subroto Sarma,Begum Fatema Zohara*. Identification, Molecular Confirmation, and Antibiotic Sensitivity of Bacteria Isolated from Repeat Breeder Cows in Rangpur Division. Insights Vet Sci. 2025: doi: 10.29328/journal.ivs.1001047; 9: 008-17
Most Viewed
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Evaluation of In vitro and Ex vivo Models for Studying the Effectiveness of Vaginal Drug Systems in Controlling Microbe Infections: A Systematic ReviewMohammad Hossein Karami*, Majid Abdouss*, Mandana Karami. Evaluation of In vitro and Ex vivo Models for Studying the Effectiveness of Vaginal Drug Systems in Controlling Microbe Infections: A Systematic Review. Clin J Obstet Gynecol. 2023 doi: 10.29328/journal.cjog.1001151; 6: 201-215
-
Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian RandomizationYong-Qing Zhu, Xiao-Yan Meng, Jing-Hua Yang*. Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian Randomization. Arch Asthma Allergy Immunol. 2023 doi: 10.29328/journal.aaai.1001032; 7: 012-022
-
Impact of Latex Sensitization on Asthma and Rhinitis Progression: A Study at Abidjan-Cocody University Hospital - Côte d’Ivoire (Progression of Asthma and Rhinitis related to Latex Sensitization)Dasse Sery Romuald*, KL Siransy, N Koffi, RO Yeboah, EK Nguessan, HA Adou, VP Goran-Kouacou, AU Assi, JY Seri, S Moussa, D Oura, CL Memel, H Koya, E Atoukoula. Impact of Latex Sensitization on Asthma and Rhinitis Progression: A Study at Abidjan-Cocody University Hospital - Côte d’Ivoire (Progression of Asthma and Rhinitis related to Latex Sensitization). Arch Asthma Allergy Immunol. 2024 doi: 10.29328/journal.aaai.1001035; 8: 007-012
-
An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergiesNathalie Cottel,Aïcha Dieme,Véronique Orcel,Yannick Chantran,Mélisande Bourgoin-Heck,Jocelyne Just. An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergies. Arch Asthma Allergy Immunol. 2021 doi: 10.29328/journal.aaai.1001027; 5: 030-037

If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."