More Information
Submitted: October 30, 2023 | Approved: November 17, 2023 | Published: November 20, 2023
How to cite this article: Mbaye F, Watt O, Sembene M. Association of HLA-DRB1 Alleles with Rheumatic Fever Among Senegalese Patients. Ann Proteom Bioinform. 2023; 7: 021-028.
DOI: 10.29328/journal.apb.1001022
Copyright License: © 2023 Mbaye F, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Rheumatic fever; Mutations; HLA-DRB1
Association of HLA-DRB1 Alleles with Rheumatic Fever Among Senegalese Patients
Fatimata Mbaye*, Oumar Watt and Mbacké Sembene
Genomics Laboratory, Department of Animal Biology, Faculty of Science and Technology, Cheikh Anta Diop University of Dakar, Dakar, BP 5005, Senegal
*Address for Correspondence: Fatimata Mbaye, Genomics Laboratory, Department of Animal Biology, Faculty of Science and Technology, Cheikh Anta Diop University of Dakar, Dakar, BP 5005, Senegal, Email: [email protected]
Background: Acute rheumatic fever (ARF) is a systemic inflammatory disease resulting from an abnormal immune response to group A β-hemolytic streptococci. ARF is a major public health problem in developing countries, particularly in Senegal. The aim of this study was to evaluate the mutation penetrance and genetic diversity of exon 2 of the HLA-DRB1 gene in Senegalese patients with ARF.
Results: DNA was extracted from the blood of patients with ARF. Exon 2 of the HLA-DRB1 gene was amplified by polymerase chain reaction and sequenced using the Sanger method. Bioinformatics software and databases (polyphen-2, SIFT and ProVean) were used to assess the pathogenicity of missense mutations. The results revealed a high level of polymorphism in exon 2 of the HLA-DRB1 gene, with 73 non-synonymous mutations between codons 21 and 89, which lie in the hypervariable region encoded by exon 2. Of the 73 variants tested, 44% were pathogenic, indicating their potential involvement in ARF onset.
Conclusion: Our results indicate that the HLA-DRB1 mutations involvement in the onset of rheumatic fever.
The immune system is part of the body’s defense system, protecting against viral and bacterial infections, parasitic infestations and other physical and chemical assaults on the body. The immune system identifies and attacks foreign antigens. However, in some cases, this system can fail and attack self-antigens, resulting in autoimmune diseases such as rheumatoid arthritis, multiple sclerosis, Graves’ disease, Bouillaud’s disease and Acute Rheumatic Fever (ARF). The latter occurs as a result of an inappropriate immune response against infection of group A β-hemolytic streptococcus (Streptococcus pyogenes) [1,2]. ARF is a post-infectious, non-suppurative sequela of pharyngeal infection with group A β-hemolytic Streptococcus [3]. It is generally characterized by arthritis (75%), fever (90%), carditis (50% - 70%), chorea (30%), and subcutaneous nodules (0% - 10%) [4].
ARF is a predominantly pediatric disease [2,4-6]; therefore, its incidence is the highest in children aged 10 years - 14 years, varying between 300,000 and 350,000 new cases annually [2]. ARF affects between approximately 15.6 and 19.6 million people worldwide [5]. The 2018 World Health Organization (WHO) report noted that 30 million people were estimated to have Chronic Rheumatic Heart Disease (CRC), which was responsible for 305,000 deaths and 11.5 million disabilities in 2015. Developing countries, particularly those in sub-Saharan Africa, Southeast Asia, and the western Pacific region, are the regions most affected by CRC, with a total prevalence rate of 84% and mortality rate of 80% [6]. The estimated economic loss caused by CRC-related deaths is US $2200 billion [6]. ARF and its sequela CRC constitute a major public health problem, particularly in sub-Saharan Africa, where the incidence is estimated at 5/1000 [7]; according to Mirabel, et al. [8], the highest figures reach 50/1000. The prevalence of acute articular rheumatism in Senegal is 30.5% [9]. Rheumatic heart disease is the leading cause of cardiovascular pathologies in pediatrics [10].
Despite the various strategies employed to combat this disease, namely primary prevention and secondary prophylaxis, ARF remains a challenge in developing countries [8]. This has garnered the attention of several researchers, some of whom have associated this disease with various risk factors, including poor housing and health conditions [2,5,11]. In addition, family aggregation and similarity in disease signs between twins and siblings suggest a genetic susceptibility to this pathology [3,12]. Among the various genes associated with ARF, the most common and well-known are those of the Human Leukocyte Antigen (HLA) system. According to Guilherme, et al. [11] and Carapetis, et al. [2], polymorphisms in most genes that encode immune system proteins are associated with ARF and CRC. Therefore, we hypothesized that mutations in certain HLA system genes predispose individuals to ARF. However, no study on this subject has yet to be published in relation to Senegal. Therefore, the aim of this study was to evaluate the penetrance, genetic diversity and evolution of exon 2 of the HLA-DRB1 gene in Senegalese patients with ARF.
Study population
Sampling was conducted in patients with ARF followed at the Clinique de Chirurgie Thoracique et Cardiovasculaire [Thoracic and Cardiovascular Surgery Clinic] of the Centre Hospitalier National Universitaire de Fann [Fann National Teaching Hospital] in Dakar, Senegal. This study was approved by the Institutional Review Board of the Cheikh Anta Diop University in Dakar (Reference No.: Protocol 0086/2015/CER/UCAD). Peripheral blood was collected in ethylenediaminetetraacetic acid tubes from individuals diagnosed with the disease and from healthy control.
DNA extraction, amplification and sequencing of exon 2 of the HLA-DRB1 gene
Total DNA was extracted from the blood of all individuals using a Zymo Research Blood Kit (Quick-DNA) according to the manufacturer’s protocol. Exon 2 of the HLA-DRB1 gene was amplified using polymerase chain reaction (PCR) in a 50 μl reaction volume containing 2 μl of concentrated DNA and 48 μl of PCR mix with 29.8 μl of MilliQ water, 5 μl of buffer, 1 μl of additional MgCl2, 2 μl of dNTPs (dATP, dCTP, dGTP, and dTTP), 5 μl of DRB forward primer, 5 μl of DRB reverse primer, and 0.2 μl of Taq polymerase. The selected sense (5′GTTCGTGTCCCCACAGCACGTTTC-3′) and anti-sense (5′-CATGCTCACCTCGCCGCTGCAC-3′) primers start in intronic regions, enabling the amplification of the whole exon 2 (270 bp). The program included an initial denaturation at 95 °C for 5 min; followed by 40 cycles of denaturation at 96 °C for 1 min, hybridization at 55 °C for 1 min, and elongation at 72 °C for 1 min 30 s; and a final extension at 70 °C for 10 min. Sequencing reactions were conducted using an MJ Research PTC-225 Peltier thermocycler with an ABI PRISM Kit, and the PCR products were subjected to electrophoresis using an ABI 3730 XL sequencer.
Chromatogram analysis
Post-sequencing chromatograms were analyzed using Mutation Surveyor software version 5.0.1 (www.softgenetics.com) to determine the position, nature, status, and identification of each mutation. The aim was to compare the sequences of individuals with the disease with the reference sequences in a software database. The software shows the position of each mutation and provides a score that indicates the probability that the detected mutation is real. A mutation is considered real only if it has a Phred score ≥20 with an accuracy score greater than 99%.
Non-synonymous mutation identification
Post-sequencing chromatograms were processed using BioEdit Version 7.2.5 software [13]. After processing and aligning the sequences, all the mutations leading to amino acid substitutions were identified. Using MEGA 7 version 7.0.14 [14], the nucleotide sequences were converted into amino acid sequences with a third reading frame that did not contain stop codons. This exercise identified the codon numbers with non-synonymous mutations and their corresponding nucleotides. We used BioEdit Version 7.2.5 [13] to identify nucleotide mutations that led to amino acid changes at each codon.
We performed BLASTP searches in GenBank to identify amino acid positions in our sequences for pathogenicity testing.
Pathogenicity test for non-synonymous mutations
Various software programs such as SIFT (http://sift.jcvi.org/) [15], Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/) [16], and Provean (http://provean.jcvi.org/) [17] were used to determine the likely pathogenic effects of non-synonymous mutations. These different types of programs act at different levels and provide different interpretation scores that indicate whether the mutation in question has had an impact on the nature of the protein, with a higher likelihood of leading to a disturbance in metabolic function.
Allele matching analysis
We performed sequence alignments using the IPD-IMG/HLA database to check for probable similarities between our sequences and the officially named HLA alleles (https://www.ebi.ac.uk/ipd/imgt/hla/). This specialized database includes official sequences named by the WHO Nomenclature Committee for HLA factors.
Genetic diversity assessment
DnaSP software version 5.10[18] was used for genetic diversity analyses to obtain the total number of sites (n), number of variable and invariable sites, variable sites of informative and non-informative types, total number of mutations (Eta), number of haplotypes (h) [19], and average number of nucleotide differences (k) [20]. MEGA 7 software version 7.0.14 [14] was used to determine the mutation rate (R).
Selection tests
A codon selection test was used to identify the evolution of mutations in exon 2 of the HLA-DRB1 coding gene. The codon selection test (dN-dS) was performed using MEGA 7 version 7.0.14 [14] with the assumption of neutral evolution (dN=dS), with a positive value indicating an overabundance of non-synonymous mutations. Thus, a P-value representing the probability of rejecting the null hypothesis was calculated.
Mutation identification
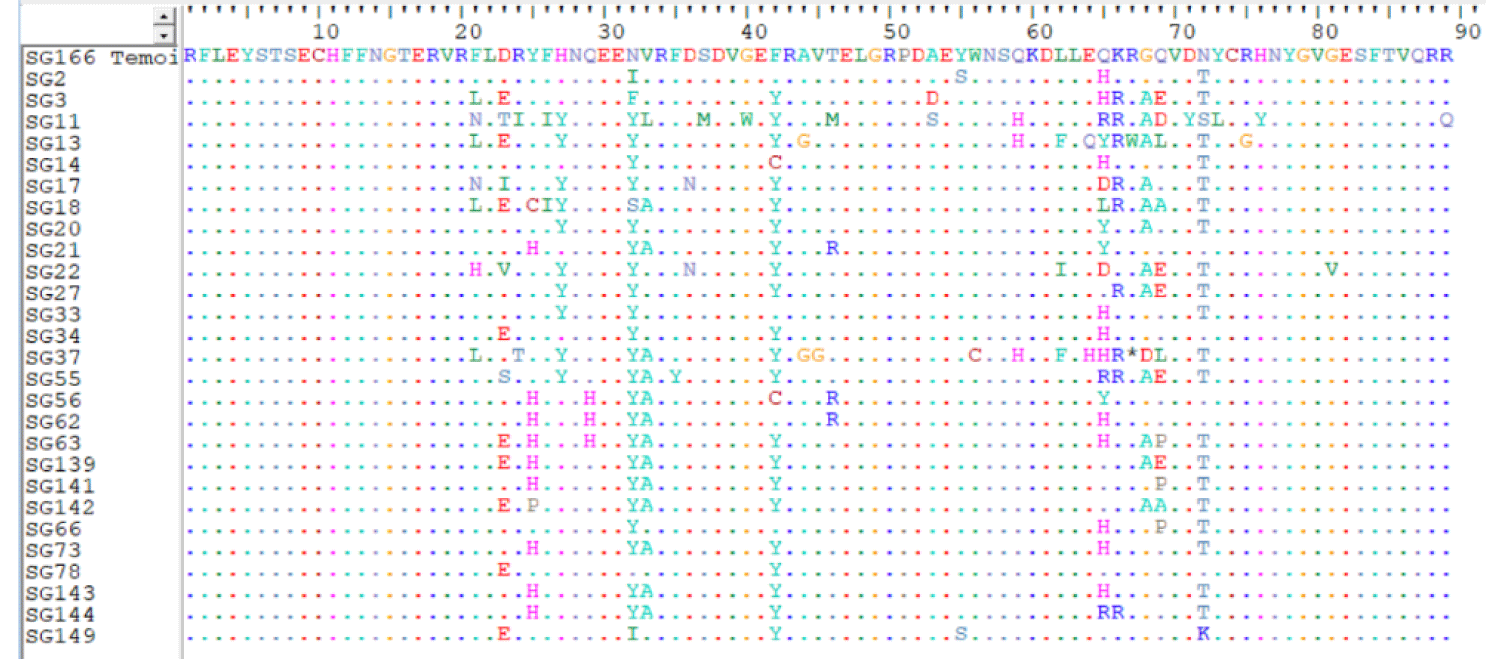
A comparison of our sequences with the Mutation Surveyor reference sequences revealed several mutations. Figure 1 summarizes the amino acid changes in the individuals compared to the control sequence (SG66), indicating the conservation of the sequences at the beginning and end. Amino acid changes began at codon 21 and the majority remained in the middle of the sequence between codons 21 and 72.
Figure 1: Multiple alignments of amino acid sequences in exon 2 of the HLA-DRB1 gene.
Mutation properties
A total of 73 mutational points leading to amino acid changes were found at 34 sites, confirming significant nucleotide polymorphisms in exon 2 of the HLA-DRB1 gene. This level of polymorphism is remarkable because some codons change by two or three bases, indicating that the entire codon can be mutated. For example, codon 23 exhibited a GAC>ACA change, leading to a change from aspartic acid to isoleucine (23.D>I). Two base changes were noted for the same codon, GAC>AGC and GAC>ACC, which led to serine (23.D>S) and threonine (23.D>T), respectively. These two base changes were also found in other codons, namely codon 64 (GAG>CAC) with a change from glutamic acid to histidine (64. E>H), codon 65 (CAG>GAC) with a change from glutamine to aspartic acid (65. Q>D), codon 69 (CAG>GCG) with a change from glutamine to aspartic acid (69.Q>D), codon 72 (AAC>CTC) with a change from asparagine to serine (72.N>S), and codon 73 (TAC>TCA) with a change from leucine to tyrosine (73.L>Y). We also found several amino acid changes within a single codon. This was the case for codons 23, 65, and 69 with five amino acid changes; codons 21, 25, 32, and 72 with three amino acid changes; and codons 24, 33, 42, 46, 53, 62, 64, and 68 with two amino acid changes.
Pathogenicity testing of the non-synonymous mutations revealed that 44% of the mutations were pathogenic. Among the codons affected by the mutations tested as pathogenic, codons 42 and 65 were the most variable in the population, with percentages of 85.18% and 77%, respectively. The others represented relatively medium or low percentages of the population. Some codons varied within a single individual with a single mutation. This was the case for codons 29 (ACG>CAT), 36 (GAC>AAC), 37 (AGC>ATC), 40 (GGG>TGG), 44 (GCG>GGG), 46 (ACG>AGG), 55 (TAC>TCC), 59 (CAG>CAC), 67 (CGG>TGG), 71 (GAC>TAC), 75 (AGA> GGA), and 89 (CGA> CAA). Codons 32 and 72 showed varying degrees of variability, with values of 96.29% and 81.48%, respectively, although the amino acid changes in these codons were non-pathogenic.
Of the mutations leading to amino acid changes, 81.94% occurred in the first two bases and 18.05% occurred at the third base. Transversion mutations significantly outnumbered transition mutations with rates of 62.82% and 37.18%, respectively (Table 1).
| Table 1: Properties of coding single nucleotide polymorphisms. | ||||||||
| Variants No. | Position on exon 2 HLA-DBR1 |
Position on HLA-DRB1 | Codon change | Nature | Mutation predictions | Findings | ||
| Provean | SIFT | Polyphen-2 | ||||||
| 1 | F21N | 55 | TTC>ATC | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 2 | F21L | 55 | TTC>TTG | T | Noxious | Tolerable | Benign | Benign |
| 3 | F21H | 55 | TTC>CTC | t | Noxious | Harmful | Prob. Harmful | Pathogen |
| 4 | D23E | 57 | GAC>GAA | T | Neutral | Tolerable | Benign | Benign |
| 5 | D23S | 57 | GAC>AGC | tt | Neutral | Harmful | Benign | Benign |
| 6 | D23I | 57 | GAC>ATA | tTT | Noxious | Tolerable | Benign | Benign |
| 7 | D23T | 57 | GAC>ACC | tT | Neutral | Harmful | Benign | Benign |
| 8 | D23V | 57 | GAC> GTC | T | Noxious | Harmful | Benign | Pathogen |
| 9 | R24I | 58 | AGA>ATA | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 10 | R24T | 58 | AGA>ACA | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 11 | Y25C | 59 | TAC>TGC | t | Noxious | Harmful | Benign | Pathogen |
| 12 | Y25H | 59 | TAC>TAT | t | Neutral | Tolerable | Benign | Benign |
| 13 | Y25P | 59 | TAC>TCC | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 14 | F26I | 60 | TTC>ATC | T | Neutral | Tolerable | Benign | Benign |
| 15 | H27Y | 61 | CAT>TAT | t | Neutral | Tolerable | Benign | Benign |
| 16 | Q29H | 63 | CAG>CAT | T | Noxious | Harmful | Benign | Pathogen |
| 17 | N32Y | 66 | AAC>TAC | T | Neutral | Tolerable | Benign | Benign |
| 18 | N32I | 66 | AAC>ATC | T | Neutral | Tolerable | Benign | Benign |
| 19 | N32S | 66 | AAC>ACT | T | Neutral | Tolerable | Benign | Benign |
| 20 | V33L | 67 | GTG>GCG | t | Neutral | Tolerable | Benign | Benign |
| 21 | V33A | 67 | GTG>TTG | T | Neutral | Tolerable | Benign | Benign |
| 22 | F35Y | 69 | TTC>TAC | T | Noxious | Tolerable | Benign | Benign |
| 23 | D36N | 70 | GAC>AAC | t | Noxious | Harmful | Benign | Pathogen |
| 24 | S37M | 71 | AGC>ATC | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 25 | G40W | 74 | GGG>TGG | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 26 | F42Y | 76 | TTC>TAC | T | Neutral | Tolerable | Benign | Benign |
| 27 | F42C | 76 | TTC>TGC | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 28 | A44G | 78 | GCG>GGG | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 29 | T46R | 80 | ACG>AGG | T | Noxious | Harmful | Poss. Harmful | Pathogen |
| 30 | T46M | 80 | ACG>ATG | t | Noxious | Harmful | Prob. Harmful | Pathogen |
| 31 | A53S | 87 | GCC>TCC | t | Noxious | Harmful | Prob. Harmful | Pathogen |
| 32 | A53D | 87 | GCC>GTC | t | Noxious | Harmful | Prob. Harmful | Pathogen |
| 33 | Y55S | 89 | TAC>TCC | T | Noxious | Tolerable | Benign | Pathogen |
| 34 | W56C | 90 | TGG>TGC | T | Noxious | Harmful | Poss. Harmful | Pathogen |
| 35 | Q59H | 93 | CAG>CAC | T | Noxious | Harmful | Benign | Pathogen |
| 36 | L62F | 96 | CTC>TTC | t | Neutral | Tolerable | Benign | Benign |
| 37 | L62I | 96 | CTC>ATC | T | Neutral | Tolerable | Benign | Benign |
| 38 | E64Q | 98 | GAG>CAG | T | Noxious | Harmful | Benign | Benign |
| 39 | E64H | 98 | GAG>CAC | TT | Noxious | Harmful | Prob. Harmful | Pathogen |
| 40 | Q65H | 99 | CAG>CAC | T | Neutral | Harmful | Benign | Benign |
| 41 | Q65R | 99 | CAG>CGG | t | Neutral | Tolerable | Benign | Benign |
| 42 | Q65L | 99 | CAG>CTG | T | Noxious | Harmful | Benign | Pathogen |
| 43 | Q65Y | 99 | CAG>TAG | t | Noxious | Harmful | Benign | Pathogen |
| 44 | Q65D | 99 | CAG>GAC | TT | Neutral | Tolerable | Benign | Benign |
| 45 | K66R | 100 | AAG>AGG | t | Neutral | Tolerable | Benign | Benign |
| 47 | R67W | 101 | CGG>TGG | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 48 | G68A | 102 | GGC>GCC | T | Neutral | Tolerable | Benign | Benign |
| 49 | G68D | 102 | GGC>GAC | t | Neutral | Harmful | Benign | Benign |
| 50 | Q69E | 103 | CAG>GAG | t | ||||
| 51 | Q69L | 103 | CAG>CTG | T | ||||
| 52 | Q69A | 103 | CAG>GCG | TT | ||||
| 53 | Q69P | 103 | CAG>CCG | T | ||||
| 54 | Q69D | 103 | CAG>GAC | TT | ||||
| 55 | D71Y | 105 | GAC>TAC | T | Noxious | Harmful | Prob. Harmful | Pathogen |
| 56 | N72T | 106 | AAC>ACC | T | Neutral | Tolerable | Benign | Benign |
| 57 | N72S | 106 | AAC>CTC | Tt | Neutral | Harmful | Benign | Benign |
| 58 | N72K | 106 | AAC>AAA | T | Neutral | Harmful | Benign | Benign |
| 59 | Y73L | 107 | TAC>TCA | TT | Noxious | Harmful | Benign | Pathogen |
| 60 | R75G | 109 | AGA>GGA | t | Noxious | Harmful | Prob. Harmful | Pathogen |
| 61 | H76Y | 110 | CAC>TAC | t | Noxious | Tolerable | Benign | Benign |
| 62 | G81V | 115 | GGG>GTG | T | Benign | Benign | ||
| 63 | R89Q | 123 | CGA>CAA | t | Noxious | Harmful | Benign | Pathogen |
| Prob. Harmful = Probably Harmful; Poss. Harmful = Possibly Harmful; T = Transversion; t = Transition | ||||||||
Matches between HLA-DRB1 alleles and patient sequences
Alignment of our sequences in the IPD-IMG/HLA database revealed several similarities. Table 2 summarizes the correspondence between our sequences and the HLA-DRB1 alleles officially designated by the WHO Nomenclature Committee, with similarity rates ranging from 90.4% to 99.6%. The results revealed a higher or lower percentage of alleles belonging to the HLA-DRB1*03 group, representing alleles coding for the DR3 antigen. Notably, the control sequence showed 99% similarity to the HLA-DRB1*03:178 allele, which is part of the HLA-DRB1*03 allele group. The HLA-DRB1*14 allele group was second at 21.42%. Sequences SG37 and SG13 corresponded to the HLA-DRB1*08 allele group, whereas sequences SG17, SG20, and SG33 corresponded to the HLA-DRB1*D10086659 allele group. These two sequences correspond to the HLA-DRB1*13MV0805 allele. Sequences SG55 and SG62 were assigned the US patent numbers 2790 and 2524, respectively.
| Table 2: Matches between HLA-DRB alleles and patient sequences. | ||
| Individual | Matching allele | Similarity rate |
| Indicator | HLA-DRB1*03*178 | 99% |
| Sg2 | New HLA-DRB1*03 | 97.8% |
| Sg3 | HLA-DRB1*14:20 | 97.4% |
| Sg11 | HLA-DRB1*1414 | 90.4% |
| Sg13 | HLA-DRB1*08 | 96.3% |
| Sg14 | New HLA-DRB1*03 | 97.8% |
| Sg17 | HLA-DBR1*D10086659 | 96.3% |
| Sg18 | Exon 2 HLA-DBR1 | 98.9% |
| Sg20 | HLA-DBR1*D10086659 | 98.5% |
| Sg21 | Exon 2 HLA-DRB1 | 98.9% |
| Sg22 | HLA-DRB1*13MV0805 | 96.7% |
| Sg27 | Exon 2 HLA-DRB1 | 98.9% |
| Sg33 | HLA-DBR1*D10086659 | 98.1% |
| Sg34 | HLA-DRB1*0327 | 98.9% |
| Sg37 | HLA-DRB1*08 | 94.8% |
| Sg55 | 2790 of the US patent | 97,4 |
| Sg56 | HLA-DRB1*13KM | 96.7 |
| Sg62 | 2524 of the US patent | 97% |
| Sg63 | HLA-DRB1*14 :02 variant | 97% |
| Sg66 | Exon 2 HLA-DRB1 | 97.8 |
| Sg73 | HLA-DRB1*0340 | 97% |
| Sg78 | HLA-DRB1*0327 | 99.6% |
| Sg139 | HLA-DRB1*14 :06 :01 | 97% |
| Sg141 | HLA-DRB1*0340 | 97.8% |
| Sg142 | HLA-DRB1*14 :02 variant | 97,4% |
| Sg143 | New HLA-DRB1*03 | 97% |
| Sg144 | HLA-DRB1*1414 | 97.8% |
| Sg149 | HLA-DRB1*0327 | 98.5 |
Diversity parameters of exon 2 of the HLA-DRB1 gene
Table 3 summarizes the genetic diversity parameters of exon 2 of the HLA-DRB1 gene in the study population. Of the 270 sites analyzed, 211 were consistent and 59 were variable (21.18%), including 25 singleton and 34 informative variable sites. These informative variabilities are likely to lead to a change in amino acids at the site and consequently to a change in protein conformation and function. A total of 78 mutations were identified. The number of haplotypes matching the number of sequences indicated that all sequences differed despite originating from individuals with ARF. The nucleotide diversity (Pi) and the average number of nucleotide differences (k) reflect the varying degrees of nucleotide variability. A mutation rate of R = 1.04 was found.
| Table 3: Genetic diversity parameters of the HLA-DRB1 gene. | |
| Genetic diversity parameters | Values |
| Population size | 28 |
| Total number of sites | 270 |
| Non-variable sites | 211 |
| Variable sites | 59 |
| Singleton-type variable sites | 25 |
| Informative-type variable sites | 34 |
| Total number of mutations | 78 |
| Number of haplotypes | 28 |
| Haplotypic diversity (Hd) | 1 |
| Nucleotide diversity (Pi) | 0.04766 |
| Average number of nucleotide differences (k) | 12.868 |
| Mutation rate ® | 1.04 |
Selection test
The codon selection test indicated an overabundance of non-synonymous mutations with a statistical value of 0.918. The test was conducted under the assumption of neutral outcomes. The probability value (prob = 0.103) indicated a positive outcome.
This study aimed to identify molecular biomarkers that could facilitate diagnostic and treatment decision-making in ARF. This was achieved using mutational analysis, estimation of genetic variability and analysis of the progress of exon 2 of the HLA-DRB1 gene. The selection of this exon was driven by its extreme polymorphism and role in immune responses. HLA-DRB1 is the most polymorphic gene in the human genome and is responsible for encoding antigen recognition peptides [21,22]. The present study included individuals with ARF who were admitted to the Center Hospitalier National Universitaire de Fann.
The results showed a high rate of nucleotide polymorphisms with 78 mutations, 73 of which resulted in amino acid changes. This high degree of polymorphism can be explained by the role of the proteins encoded by HLA class II genes, especially exon 2 of the HLA-DRB1 gene. The latter encodes antigen-presenting peptides for CD4+ T cells and B lymphocytes in specific immune response processes [23]. This antigen recognition process leads to clonal selection of lymphocytes capable of producing antibodies for a given response. Lymphocytes are often required to increase or alter their antigenic or immunoglobulin repertoires. This phenomenon occurs via site-specific recombination. Salomé & Kukkonen [23] identified a signal sequence potentially involved in recombination in exon 2, from nucleotide 254 to nucleotide 260 (5′-CACGGTG-3′). The present study also showed that the first three nucleotides (CAC) are the most influential in recombination, with the fourth nucleotide mutated in only 7% of alleles. This sequence was present in our population with a transitional mutation in the fourth nucleotide (G>A). These recombinations have contributed to an increase in the number of alleles of the HLA-DRB1 gene. In 2008, 540 HLA-DRB1 alleles were identified, making it the most polymorphic locus in the human genome [23]. Wysocki, et al. [22] reported 2690 HLA-DRB1 alleles in 2020. Additionally, a recombination-promoting region was found in all alleles of the same exon between positions 145 and 150, except for the HLA-DRB1*07 allele [23]. This extreme polymorphism was also manifested at the codon level. Some of these proteins are affected by amino acid changes. Codons 23 (eight changes) and 65 (six changes) remained the most affected. Furthermore, three mutations were involved in the conversion of aspartic acid to isoleucine (GAC>ATA) at codon 23.
The 73 mutations leading to amino acid changes remained concentrated at 34 sites, representing a ratio of 2.14 mutations per site. These mutations affect codons 21 - 89 of exon 2, which correspond to codons 55 - 123 of the complete HLA-DRB1 gene. The portion between codons 21 and 89 contains a hypervariable region encoded by exon 2. These three hypervariable regions are the most prone to amino acid changes [22]. The third region (encoded by exon 2 of the HLA-DRB1 gene) lies between amino acids 67 to 74 in the alpha helix of the β1 chain, which is among the most important sites in antigen recognition in CD4+ T cells. Mutations in this hypervariable region can lead to a change in the antigenic specificity of CD4+ T cells, causing them to recognize self-antigens, thus inducing autoimmune responses. This was also discussed by Wysocki, et al. [22], who reported that mutations in this hypervariable region would lead to alleles that cause cross-reactivity, resulting in recognition of self-antigens by lymphocytes. This phenomenon is recognized as the process by which autoimmune diseases such as ARF arise. Wysocki, et al. [22] also identified three amino acid sequences (70QRRAA74, 70RRRAA74, and 70QKRAA74) that may be responsible for the production of self-antigens by T cells. These sequences are associated with rheumatoid arthritis [22]. Although absent in our sequences, they can be considered evidence of possible antigenic similarity (foreign and self-antigens) due to amino acid changes and the likely existence of other sequences involved in epitope similarities.
In our sequences, this hypervariable region encoded by exon 2 of the HLA-DRB1 gene corresponds to amino acids 33 - 40. Five of these residues were changed: valine at position 33, phenylalanine at position 35, aspartic acid at position 36, serine at position 37, and glycine at position 40. Changes from aspartic acid to asparagine, serine to methionine, and glycine to tryptophan at positions 36, 37, and 40, respectively, were tested for pathogenicity. Thus, this region could be considered a target for researching molecular markers of autoimmune diseases, including ARF.
Pathogenicity tests indicated that 44% of mutations leading to amino acid changes may be involved in the onset of ARF in affected individuals. This could be explained by the fact that these amino acid changes affect protein conformation, function, and specificity. A protein involved in lymphocyte antigen recognition can cross-react with foreign antigens (S. pyogenes antigens) and self-antigens, and may be affected by amino acid changes that affect its function. Changes in the hypervariable regions may also explain these pathogenicities. Codons 42 and 65 stand out from the crowd owing to their high variability in the population with a frequency of 85.18% and 77%, respectively. Notably, codon 42 was located immediately after the variable region and codon 65 was the most affected by amino acid changes. This high variability was also observed for codons 32 and 72. However, these were not tested for pathogenicity. These four codons (32, 42, 65 and 72) could also be considered leads for researching ARF molecular markers.
The modification in the first two codon bases consistently had the most remarkable impact on amino acid changes, with 81.94% versus 18.06% changes in the third base. This can be attributed to the redundancy of the genetic code, indicating that changes in the first two bases induce more amino acid changes. Moreover, changes in the third base often have no effect at the amino acid level. However, in some cases, these mutations can induce changes, as observed in 18.06% of the mutations in our population. The preponderance of transitional mutations in our sequences was consistent with that reported by Salomé & Kukkonen [23]. This previous study, which investigated all exons (1-6) of the HLA-DRB1 gene, found a transitional mutation rate in exon 2 that far exceeded those of the other exons combined.
Despite the high rate of polymorphisms in our sequences, codons 1-22 were not affected by these mutations. These codons corresponded to the first 62 nucleotides of the sequences. This sequence consistency was also found in the study by Salomé and Kukkonen [23], which revealed the conservation of nucleotides from 3 to 13.
Alignment results from the IPD-IMG/HLA database showed that 17.85% of our sequences resembled alleles in the HLA-DRB1*03 group. This percentage is similar to that of the HLA-DRB1*14 allele group. Other similarities with HLA-DRB1 alleles, namely HLA-DRB1*08 alleles, were also found. These similarities can be explained by the fact that these alleles contribute to the breakdown of immune tolerance, leading to reactions directed against self-antigens. The HLA-DRB1*03 allele is associated with Lögren’s syndrome. This disease presents with symptoms similar to those of ARF, including fever, arthritis, and erythema. The HLA-DRB1 *08 allele is found in the HLA-DR8/17 haplotype [3]. This study showed that the HLA-DR8/17 haplotype was most frequently associated with acute articular rheumatism. However, no similarity was observed with the HLA-DRB1*07 allele, which is the most implicated allele in acute articular rheumatism. This could be attributed to the population-based diversity of HLA system genes. Thus, the HLA system has population-based allelic specificity worldwide. Hamed, et al. [24] found the HLA-DRB1*03 allele in 24.73% of 30 light-skinned moors, 34 dark-skinned moors, and 29 Black Africans living in Mauritania. No HLA-DRB1*07 alleles were detected in this population. HLA-DRB1* 08:06, DRB1:08:04:01 alleles were found at high frequencies in sub-Saharan African, North African, and Black European populations in a study by Vina, et al. [25]. This study involved a sample of sub-Saharan Africans, Europeans, North Africans, and Black Europeans.
Genetic variability tests further confirmed the high polymorphism rate in exon 2 of the HLA-DRB1 gene, with 78 mutations at 59 sites, 34 of which were informative. The number of informative sites corresponds to the number of sites where amino acid changes occur. This diversity is also reflected in the number of haplotypes, which is equal to the number of individuals in the population. In other words, these two sequences were not found in our population. This is attributed to the extreme polymorphism of HLA system genes. A codon selection test confirmed the prevalence of non-synonymous mutations with a positive outcome. This is consistent with the high rate of amino acid change.
This study aimed to identify mutations in exon 2 of the HLA-DRB1 gene involved in the onset of ARF in Senegalese patients. This was achieved using a molecular approach involving mutational analysis, diversity assessment, and evolutionary testing. The results aligned with our initial hypothesis regarding the involvement of HLA gene mutations in ARF development. In total, 73 variants leading to amino acid changes were identified in exon 2 of the HLA-DRB1 gene. These mutations affect the structure and function of proteins.
The authors acknowledge the genomics laboratory of the department of Animal Biology, Faculty of Science and Technology, Cheikh Anta Diop University, Dakar.
- Peterson DC. Complications of physician misdiagnosis/treatment of rheumatic fever in the United States. Adv Biosci Biotechnol. 2013; 4:143-146.
- Carapetis JR, Beaton A, Cunningham MW, Guilherme L, Karthikeyan G, Mayosi BM, Sable C, Steer A, Wilson N, Wyber R, Zühlke L. Acute rheumatic fever and rheumatic heart disease. Nat Rev Dis Primers. 2016 Jan 14;2:15084. doi: 10.1038/nrdp.2015.84. PMID: 27188830; PMCID: PMC5810582.
- Seckeler MD, Hoke TR. The worldwide epidemiology of acute rheumatic fever and rheumatic heart disease. Clin Epidemiol. 2011 Feb 22;3:67-84. doi: 10.2147/CLEP.S12977. PMID: 21386976; PMCID: PMC3046187.
- Karthikeyan G, Guilherme L. Acute rheumatic fever. Lancet. 2018 Jul 14;392(10142):161-174. doi: 10.1016/S0140-6736(18)30999-1. Epub 2018 Jun 29. Erratum in: Lancet. 2018 Sep 8;392(10150):820. PMID: 30025809.
- Corsenac P, Heenan RC, Roth A, Rouchon B, Guillot N, Hoy D. An epidemiological study to assess the true incidence and prevalence of rheumatic heart disease and acute rheumatic fever in New Caledonian school children. J Paediatr Child Health. 2016 Jul;52(7):739-44. doi: 10.1111/jpc.13185. Epub 2016 May 20. Erratum in: J Paediatr Child Health. 2016 Oct;52(10 ):974. PMID: 27203400.
- World Health Organization. Rheumatic fever and rheumatic heart disease. 2018. https://www.who.int/news-room/fact-sheets/detail/rheumatic-heart-disease
- Ngaïdé AA, Mbaye A, Kane A. Prevalence of rheumatic heart disease in Senegalese schoolchildren: clinical and echocardiographic screening. Heart Asia. 2015; 7:40-45.
- Mirabel M, Ferreira B, Sidi D. Acute rheumatic fever: perspectives. Med Sci (Paris). 2012; 28:633-638.
- Diao M, Kane A, Doumbia AS. Progressive rheumatic heart disease in 17 cases collected at Dakar University Hospital. Med Trop. 2005; 65:339-342.
- Fall AL, Ndiaye O, Lavou I. Rheumatic heart disease at the Albert Royer Children’s Hospital in Dakar: about 76 cases. Conférence: Fourth Congress of the French-Speaking Black African Pediatricians Association(APANF) and Second Congress of the Senegalese Pediatric Society(SOSEPED), Dakar, Senegal. 2007.
- Guilherme L, Köhler K, Kalil J. Rheumatic heart disease: genes, inflammation and autoimmunity. Rheumatol Curr Res. 2012; 10:2161-1149.
- Azevedo PM, Pereira RR, Guilherme L. Understanding rheumatic fever. Rheumatol Int. 2012 May;32(5):1113-20. doi: 10.1007/s00296-011-2152-z. Epub 2011 Sep 28. PMID: 21953302.
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symposium Series. London. 1999; c1979-c2000:95-98.
- Tamura K, Stecher G, Humar S. MEGA 7: molecular Evolutionary Genetics Analysis. version 7.14. Mol Biol Evol. 2016; 30:2725-2729.
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001 May;11(5):863-74. doi: 10.1101/gr.176601. PMID: 11337480; PMCID: PMC311071.
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 Jan;Chapter 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. PMID: 23315928; PMCID: PMC4480630.
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7(10):e46688. doi: 10.1371/journal.pone.0046688. Epub 2012 Oct 8. PMID: 23056405; PMCID: PMC3466303.
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009 Jun 1;25(11):1451-2. doi: 10.1093/bioinformatics/btp187. Epub 2009 Apr 3. PMID: 19346325.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406-25. doi: 10.1093/oxfordjournals.molbev.a040454. PMID: 3447015.
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983; 105:437-460.
- Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet. 2013;14:301-23. doi: 10.1146/annurev-genom-091212-153455. Epub 2013 Jul 15. PMID: 23875801; PMCID: PMC4426292.
- Wysocki T, Olesińska M, Paradowska-Gorycka A. Current Understanding of an Emerging Role of HLA-DRB1 Gene in Rheumatoid Arthritis-From Research to Clinical Practice. Cells. 2020 May 2;9(5):1127. doi: 10.3390/cells9051127. PMID: 32370106; PMCID: PMC7291248.
- von Salomé J, Kukkonen JP. Sequence features of HLA-DRB1 locus define putative basis for gene conversion and point mutations. BMC Genomics. 2008 May 19;9:228. doi: 10.1186/1471-2164-9-228. PMID: 18489735; PMCID: PMC2408603.
- Hamed CT, Meiloud G, Veten F, Hadrami M, Ghaber SM, Boussaty EC, Habti N, Houmeida A. HLA class I (-A, -B, -C) and class II (-DR, -DQ) polymorphism in the Mauritanian population. BMC Med Genet. 2018 Jan 3;19(1):2. doi: 10.1186/s12881-017-0514-4. PMID: 29298671; PMCID: PMC5751816.
- Fernandez Vina MA, Hollenbach JA, Lyke KE, Sztein MB, Maiers M, Klitz W, Cano P, Mack S, Single R, Brautbar C, Israel S, Raimondi E, Khoriaty E, Inati A, Andreani M, Testi M, Moraes ME, Thomson G, Stastny P, Cao K. Tracking human migrations by the analysis of the distribution of HLA alleles, lineages and haplotypes in closed and open populations. Philos Trans R Soc Lond B Biol Sci. 2012 Mar 19;367(1590):820-9. doi: 10.1098/rstb.2011.0320. PMID: 22312049; PMCID: PMC3267126.