More Information
Submitted: April 20, 2023 | Approved: April 28, 2023 | Published: May 01, 2023
How to cite this article: Zhou A, Zhang C, Eminaga O. Advances in deep learning-based cancer outcome prediction using multi-omics data. Ann Proteom Bioinform. 2023; 7: 010-013.
DOI: 10.29328/journal.apb.1001020
Copyright License: © 2023 Zhou A, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Deep learning; Cancer; Outcome prediction; Multi-omics
Abbreviations: ACC: Accuracy; BiDNNs: Bidirectional Deep Neural Networks; C-index: Concordance Index; CNV: Copy Number Variation
Advances in deep learning-based cancer outcome prediction using multi-omics data
Andrew Zhou, Charlie Zhang and Okyaz Eminaga*
Department of Urology, Stanford University, Stanford, California 94305, USA
*Address for Correspondence: Okyaz Eminaga, Department of Urology, Stanford University, Stanford, California 94305, USA, Email: [email protected]
Cancer prognosis reflects a complex biological process measured by multiple types of omics data. Deep learning frameworks have been proposed to integrate multi-omics data and predict patient outcomes in different cancer types, potentially revolutionizing cancer prognosis with superior performance. This minireview summarizes the advances in the strategies for multi-omics data integration and the performance of different deep learning models in prognosis prediction of diverse cancer types using multi-omics data published in the past 18 months. The challenges and limitations of deep learning models for predicting cancer outcomes based on multi-omics data are discussed.
Accurate cancer prognosis prediction may necessitate the use of multiple types of omics data since a single type of omics data may not present the entire story. For example, analyzing only gene expression data may not reveal important information about protein expression or metabolite levels that could impact cancer prognosis. In contrast, the integration of multiple omics enables the construction of molecular networks that help identify holistic mechanistic pathways underlying cancer progression. This integration requires the state-of-art bioinformatic approach, i.e. deep learning-based modeling, because it can handle the high-dimensional and complex nature of the multi-omics data which involves thousands or even millions of features, and identify patterns and associations that may be missed by traditional statistical methods. Various deep learning frameworks have been proposed to integrate multi-omics data including epigenomics, genomics, proteomics, metabolomics and radiomics to predict patient outcomes in different cancer types and may revolutionize cancer prognosis with superior performance [1]. Here, we review the advances in the strategies for multi-omics data integration and the performance of different deep learning models in prognosis prediction of diverse cancer types using multi-omics data published in the past 18 months since studies reported prior to September 2021 have been reviewed elsewhere [2].
The most common types of omics data and data representations used to train deep learning models and the main types of unsupervised data integration methods for combining multi-omics data have been comprehensively reviewed [3,4]. Depending on when data integration occurs, data integration methods can also be categorized into two types, i.e., early fusion and late fusion. Studies using early fusion stack the matrices of all multi-omics data first and then construct one model to perform the prediction, while studies employing late fusion construct models for each type of multi-omics data first and then stack the information from each model for prediction. Ding et al. recently developed a supervised "cooperative learning" algorithm that combines early and late fusion methods and their blended versions to encourage the predictions using different types of omics data to agree and chooses the degree of agreement in a data-adaptive manner [5]. The powerful method allows flexible fitting mechanisms for different modalities and boosts the aligned signals across modalities by exploiting their underlying relationships, thereby, improving prediction accuracy using multi-omics data. Moreover, Benkirane, et al. developed a customizable versatile deep learning-based strategy for multi-omics integration, named CustOmics, that relies on a two-phase approach in which the training to each omics data source is adapted independently before learning cross-modality interactions in the second phase, providing highly interpretable results [6].
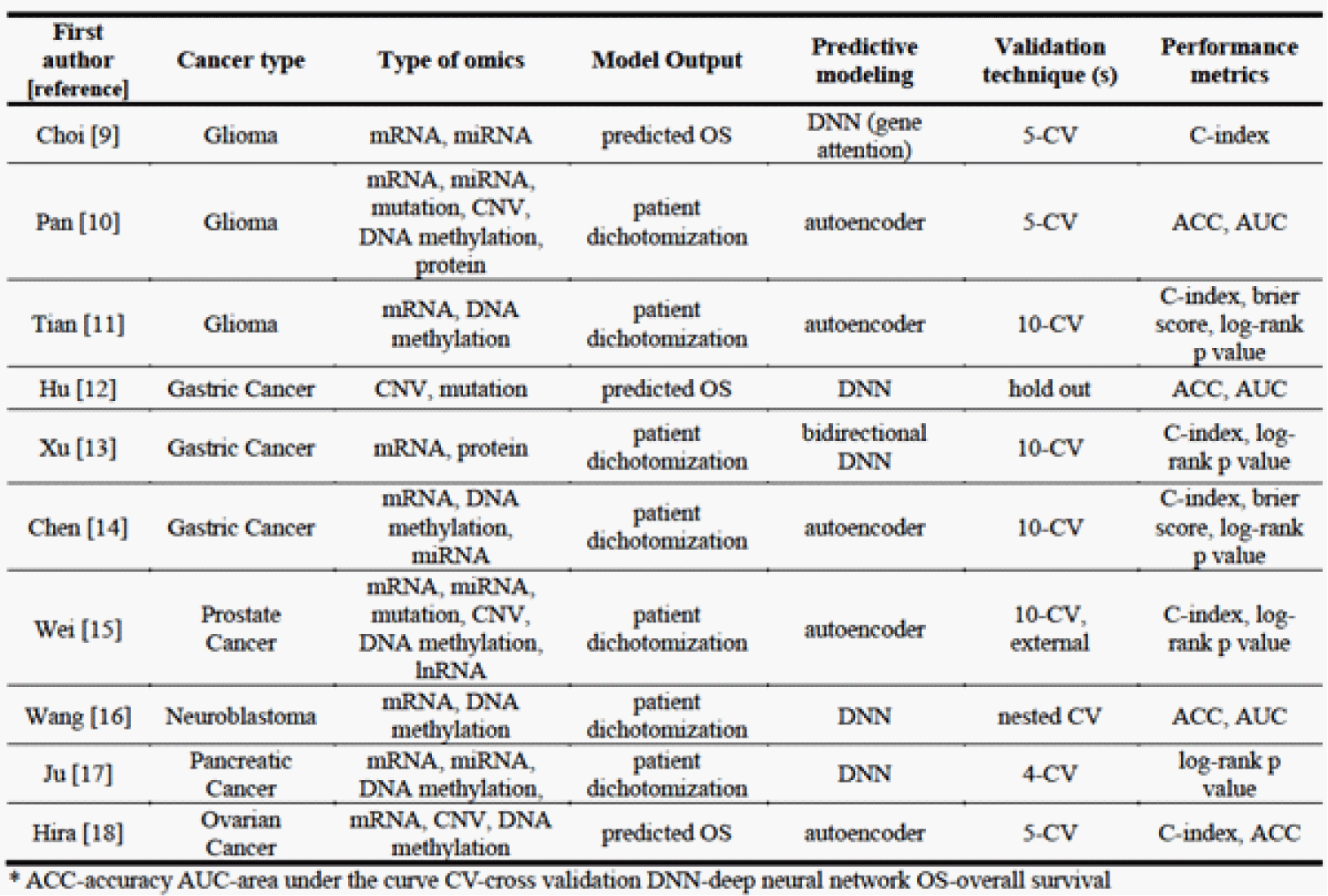
With the recent progress in deep learning techniques, various prediction models have been generated for outcome prediction using multi-omics data and their performances have been evaluated in multiple cancer types using the concordance index (c-index) [7] that measures the discriminative power of the model by comparing the predicted results with the real survival time, or accuracy (ACC) that measures how often the model correctly predicts the target variable [8]. The types of omics data used, predictive modeling method, validation technique, and performance metrics of the most recent studies published within the past 18 months were summarized in Table 1.
Table 1: Information of the most recent studies reviewed in this article.
In glioma, Multi-PEN (Multi-Prognosis Estimation Network) was developed to predict survival using mRNA and miRNA expression with a c-index of 0.70 [9], while i-Modern integrating transcription profile, miRNA expression, somatic mutations, copy number variation (CNV), DNA methylation, and protein expression achieved ACC of 97.80% when classifying glioma patients in TCGA into subgroups with differential prognosis [10]. Another deep learning model using mRNA expression and DNA methylation data achieved a c-index of 0.70-0.92 in multiple glioma datasets [11]. In gastric cancer, GCS-Net, a biological pathway-based sparse deep neural network model was recently constructed for long-term survival prediction using CNV and somatic mutation data and showed higher accuracy (c-index = 0.844) [12]. In addition, a bidirectional deep neural networks (BiDNNs) based model integrating transcriptomics and epigenomics data stratified gastric cancer patients into two survival subgroups with a c-index of 0.65 [13]. Moreover, an unsupervised feedforward neural network-based model was proposed to integrate mRNA, miRNA and methylation data and predict the prognosis of gastric cancer patients with a c-index of 0.61 to 0.71 in multiple datasets [14]. The model demonstrates better performance than the two alternative approaches to prognosis prediction. In prostate cancer, a novel deep learning-based model combining profiles of mRNA, miRNA, DNA methylation, CNVs and lncRNA predicted outcome with a c-index of 0.767 [15]. In neuroblastoma, a deep learning model using a network-level fusion of multi-omics data outperformed feature-level fusion and achieved 79% and 70% ACCs for outcome prediction on two patient cohorts [16]. In pancreatic cancer, multi-omics deep learning for prognosis-correlated subtyping (MODEL-P) was developed to integrate mRNA sequencing, microRNA sequencing, and DNA methylation data and accurately stratify patients into subgroups with distinct survival outcomes [17]. Finally, in ovarian cancer, integrating CNV, DNA methylation, and mRNA expression data using variational autoencoders in deep learning model construction showed a c-index of 0.60-0.68 in outcome prediction of multiple patient cohorts [18]. These most recent studies demonstrated the promising potential of deep learning-based models in cancer outcome prediction using multi-omics data.
Deep learning-based cancer prognosis studies using multi-omics data have demonstrated superior accuracy than traditional machine learning methods. In addition, deep learning models trained on one cancer type have been shown to be transferable to other cancer types, allowing the generalized use of a single model on multiple cancer outcome predictions. Moreover, these models helped identify novel biomarkers and important genes that contribute to patient outcomes, shedding light on the underlying mechanisms of cancer progression and therapeutic development. However, several challenges and limitations must be addressed [19,20]. First, the success of deep learning models relies on the availability and quality of data. Multi-omics data can be very complex, and obtaining large, high-quality datasets can be challenging. If the multi-omics data is noisy, incomplete, or biased, it may negatively impact the model's utility. Moreover, multi-omics data have different measurement technologies, scales, and levels of noise. The heterogeneity of the data can lead to performance degradation because the model may not effectively capture the underlying biological relationships. Second, deep learning models are known for their ability to learn complex relationships between input features and outcomes. However, special measurements are required to mitigate the model overfitting and to facilitate the model generalization on unseen data as the number of features can be very high and problematic in multi-omics data. Third, understanding the underlying biological mechanisms behind the predictions can be challenging, which limits the ability to use these models to generate new hypotheses. Fourth, deep learning models trained on multi-omics data from one population may not generalize well to other populations. This is particularly true for diseases that have different underlying genetic and environmental factors in different populations. Lastly, while deep learning models can automate many aspects of data analysis, they still require domain expertise to interpret the results correctly. Without proper domain expertise, it may be challenging to determine whether the model's predictions are biologically plausible.
As more multi-omics data becomes available, deep learning models have the potential to greatly improve survival prediction using multi-omics data, leading to better patient outcomes and advancing our understanding of complex diseases. However, it requires careful attention to data quality, model complexity, sample size and the heterogeneity of the data in order to achieve accurate, interpretable and generalizable results.
We thank Dr. Hongjuan Zhao for helpful discussions on the topic and edits of the manuscript.
- Arjmand B, Hamidpour SK, Tayanloo-Beik A, Goodarzi P, Aghayan HR, Adibi H, Larijani B. Machine Learning: A New Prospect in Multi-Omics Data Analysis of Cancer. Front Genet. 2022 Jan 27;13:824451. doi: 10.3389/fgene.2022.824451. PMID: 35154283; PMCID: PMC8829119.
- Lobato-Delgado B, Priego-Torres B, Sanchez-Morillo D. Combining Molecular, Imaging, and Clinical Data Analysis for Predicting Cancer Prognosis. Cancers (Basel). 2022 Jun 30;14(13):3215. doi: 10.3390/cancers14133215. PMID: 35804988; PMCID: PMC9265023.
- Vahabi N, Michailidis G. Unsupervised Multi-Omics Data Integration Methods: A Comprehensive Review. Front Genet. 2022 Mar 22;13:854752. doi: 10.3389/fgene.2022.854752. PMID: 35391796; PMCID: PMC8981526.
- Tsimenidis S, Vrochidou E, Papakostas GA. Omics Data and Data Representations for Deep Learning-Based Predictive Modeling. Int J Mol Sci. 2022 Oct 14;23(20):12272. doi: 10.3390/ijms232012272. PMID: 36293133; PMCID: PMC9603455.
- Ding DY, Li S, Narasimhan B, Tibshirani R. Cooperative learning for multiview analysis. Proc Natl Acad Sci U S A. 2022 Sep 20;119(38):e2202113119. doi: 10.1073/pnas.2202113119. Epub 2022 Sep 12. PMID: 36095183; PMCID: PMC9499553.
- Benkirane H, Pradat Y, Michiels S, Cournède PH. CustOmics: A versatile deep-learning based strategy for multi-omics integration. PLoS Comput Biol. 2023 Mar 6;19(3):e1010921. doi: 10.1371/journal.pcbi.1010921. PMID: 36877736; PMCID: PMC10019780.
- Uno H, Cai T, Pencina MJ, D'Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011 May 10;30(10):1105-17. doi: 10.1002/sim.4154. Epub 2011 Jan 13. PMID: 21484848; PMCID: PMC3079915.
- Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Information Processing & Management. 2009; 45:427-437. doi:https://doi.org/10.1016/j.ipm.2009.03.002
- Choi SR, Lee M. Estimating the Prognosis of Low-Grade Glioma with Gene Attention Using Multi-Omics and Multi-Modal Schemes. Biology (Basel). 2022 Oct 5;11(10):1462. doi: 10.3390/biology11101462. PMID: 36290366; PMCID: PMC9598836.
- Pan X, Burgman B, Wu E, Huang JH, Sahni N, Stephen Yi S. i-Modern: Integrated multi-omics network model identifies potential therapeutic targets in glioma by deep learning with interpretability. Comput Struct Biotechnol J. 2022 Jun 30;20:3511-3521. doi: 10.1016/j.csbj.2022.06.058. PMID: 35860408; PMCID: PMC9284388.
- Tian J, Zhu M, Ren Z, Zhao Q, Wang P, He CK, Zhang M, Peng X, Wu B, Feng R, Fu M. Deep learning algorithm reveals two prognostic subtypes in patients with gliomas. BMC Bioinformatics. 2022 Oct 11;23(1):417. doi: 10.1186/s12859-022-04970-x. PMID: 36221066; PMCID: PMC9552440.
- Hu J, Yu W, Dai Y, Liu C, Wang Y, Wu Q. A Deep Neural Network for Gastric Cancer Prognosis Prediction Based on Biological Information Pathways. J Oncol. 2022 Sep 9;2022:2965166. doi: 10.1155/2022/2965166. PMID: 36117847; PMCID: PMC9481367.
- Xu J, Yao Y, Xu B, Li Y, Su Z. Unsupervised learning of cross-modal mappings in multi-omics data for survival stratification of gastric cancer. Future Oncol. 2022 Jan;18(2):215-230. doi: 10.2217/fon-2021-1059. Epub 2021 Dec 2. PMID: 34854737.
- Chen S, Zang Y, Xu B, Lu B, Ma R, Miao P, Chen B. An Unsupervised Deep Learning-Based Model Using Multiomics Data to Predict Prognosis of Patients with Stomach Adenocarcinoma. Comput Math Methods Med. 2022 Oct 27;2022:5844846. doi: 10.1155/2022/5844846. PMID: 36339684; PMCID: PMC9633210.
- Wei Z, Han D, Zhang C, Wang S, Liu J, Chao F, Song Z, Chen G. Deep Learning-Based Multi-Omics Integration Robustly Predicts Relapse in Prostate Cancer. Front Oncol. 2022 Jun 23;12:893424. doi: 10.3389/fonc.2022.893424. PMID: 35814412; PMCID: PMC9259796.
- Wang C, Lue W, Kaalia R, Kumar P, Rajapakse JC. Network-based integration of multi-omics data for clinical outcome prediction in neuroblastoma. Sci Rep. 2022 Sep 14;12(1):15425. doi: 10.1038/s41598-022-19019-5. PMID: 36104347; PMCID: PMC9475034.
- Ju J, Wismans LV, Mustafa DAM, Reinders MJT, van Eijck CHJ, Stubbs AP, Li Y. Robust deep learning model for prognostic stratification of pancreatic ductal adenocarcinoma patients. iScience. 2021 Nov 10;24(12):103415. doi: 10.1016/j.isci.2021.103415. PMID: 34901786; PMCID: PMC8637475.
- Hira MT, Razzaque MA, Angione C, Scrivens J, Sawan S, Sarker M. Integrated multi-omics analysis of ovarian cancer using variational autoencoders. Sci Rep. 2021 Mar 18;11(1):6265. doi: 10.1038/s41598-021-85285-4. Erratum in: Sci Rep. 2021 Aug 11;11(1):16671. PMID: 33737557; PMCID: PMC7973750.
- Mathema VB, Sen P, Lamichhane S, Orešič M, Khoomrung S. Deep learning facilitates multi-data type analysis and predictive biomarker discovery in cancer precision medicine. Comput Struct Biotechnol J. 2023 Jan 31;21:1372-1382. doi: 10.1016/j.csbj.2023.01.043. PMID: 36817954; PMCID: PMC9929204.
- Nicora G, Vitali F, Dagliati A, Geifman N, Bellazzi R. Integrated Multi-Omics Analyses in Oncology: A Review of Machine Learning Methods and Tools. Front Oncol. 2020 Jun 30;10:1030. doi: 10.3389/fonc.2020.01030. PMID: 32695678; PMCID: PMC7338582.