More Information
Submitted: July 13, 2022 | Approved: July 19, 2022 | Published: July 20, 2022
How to cite this article: Al-Madhagi HA, Tahan ZS. In silico disrupting quorum sensing of porphyromonas gingivalis via essential oils and coumarin derivatives. Ann Proteom Bioinform. 2022; 6: 001-005.
DOI: 10.29328/journal.apb.1001017
Copyright License: © 2022 Al-Madhagi HA, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Quorum sensing; Molecular docking; Porphyromonas gingivalis; Essential oils; Coumarins
In silico disrupting quorum sensing of porphyromonas gingivalis via essential oils and coumarin derivatives
Haitham Ahmed Al-Madhagi1* and Zaher Samman Tahan2
and Zaher Samman Tahan2
1Division of Biochemistry, Department of Chemistry, Faculty of Science, Aleppo University, Aleppo, Syrian Arab Republic
2Division of Microbiology, Department of Plant Biology, Faculty of Science, Aleppo University, Aleppo, Syrian Arab Republic
*Address for Correspondence: Haitham Ahmed Al-Madhagi, Division of Biochemistry, Department of Chemistry, Faculty of Science, Aleppo University, Aleppo, Syrian Arab Republic, Email: [email protected]
The emergence of porphyromonas gingivalis biofilm is a hallmark of risky burden diseases including Alzheimer's disease and atherosclerosis. The current study aims to screen some natural essential oil compounds and coumarin derivatives to interfere with quorum sensing of the bacterium and thus biofilm formation. A total of 20 ligands (10 essential oil molecules and 10 coumarin derivatives) were docked to P.gingivalis heme-binding protein HmuY using UCSF Chimera built-in AutoDock interface. Alongside, ADMET properties were also predicted via ADMETsar 2.0 and ProTox-II webservers. All of the selected ligands had higher free energy values than the reference inhibitor MES and native coumarin as well. Moreover, ADME parameters are in good agreement with Lipinski's rule of five. Nevertheless, the best molecules with top binding energy exhibited slight immunogenicity as well as carcinogenicity issues requiring in vitro confirmation. In conclusion, the tested ligands had better efficacy against P.gingivalis quorum sensing and biofilm.
Numerous microbiota commensal lives in harmony within mankind's oral cavity without harming the host tissues. These bacterial communities are composed of > 700 species forming a complex, structured biofilm attached to the tongue surface or subgingival region and embedded in a thick substance produced by those bacteria. This biofilm is a heterogeneous material comprised mainly of polysaccharides and DNA [1]. If the pathogenic bacteria constitute a major part, this puts a high risk to human health since, in the biofilm form, the pathogenic bacteria become more aggressive and resistant to host immune weapons and antimicrobials administered orally [2].
One of the most commonly found bacteria within the oral cavity is porphyromonas gingivalis in addition to Tannerella forsythia and Treponema denticola (forming triad red complex) [3]. This Gram-negative, anaerobic joins the subgingival biofilm later [4]. This oral pathogen was at first thought to be associated with chronic periodontitis only. Indeed, nowadays it is well-known that P.gingivalis is linked to seemingly unpredictable types of diseases such as Alzheimer's disease (AD) [5], atherosclerosis [6], and cancers of the digestive canal [7]. This pathogenicity is mainly attributed to the high virulence pattern reflected by diverse virulence factors the bacterium uses to invade and colonize the underlying tissue plus the evading as well as interference with the immune cells' strategies [8].
Once the biofilm has been established, conventional antibiotics lack the usual effectiveness due to the polysaccharide barrier formed by the biofilm microbial community which hampers its accessibility. This prompted the creation of novel ways to deal with the pathogenic biofilm [9]. One such strategy is to target the quorum sensing (QS) pathway utilized by the bacterial community to sense the density of the species population and thus initiate biofilm formation and pathogenesis [10]. Thus, disrupting cell-cell communication eliminates unnecessary antibiotic usage and easily controls bacterial pathogenesis. This can be achieved through the use of some natural products which have proven their efficacy as antibiofilm as well as interference of the QS mechanisms [11]. Utilizing this concept, He, et al. [12] have successfully used coumarin to interfere with the QS pathway of P.gingivalis In silico and in vitro.
This prompted us to screen some essential oil compounds and coumarin derivatives to control the pathogenicity of P.gingivalis toward periodontitis and other associated chronic diseases by targeting a heme-binding protein HmuY.
Protein preparation
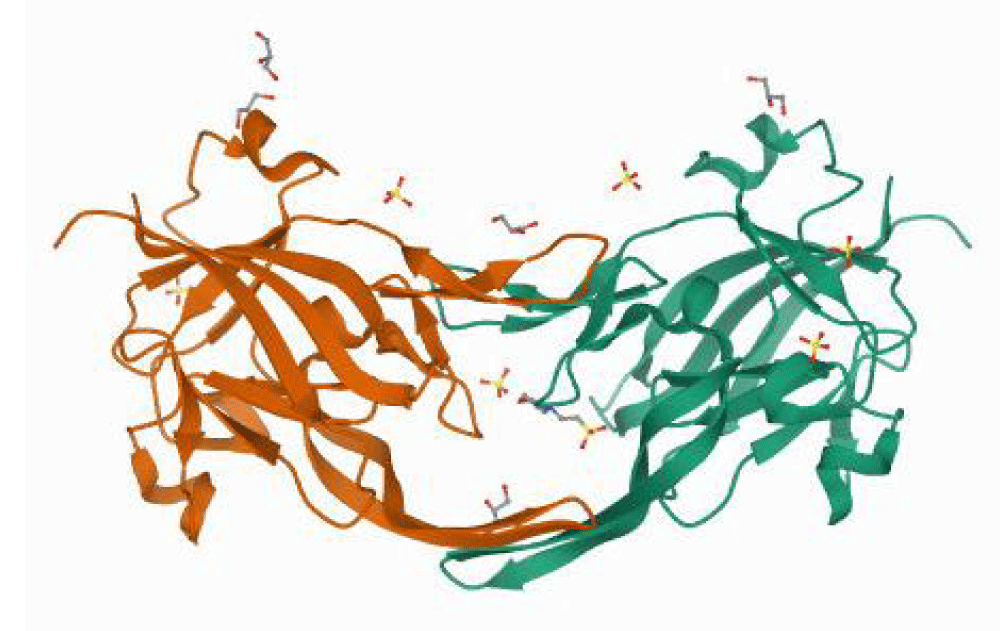
The crystal structure of the heme-free heme-binding protein of P.gingivalis HmuY complexed with 2-(n-morpholino)-ethane sulfonic acid (MES) (PDB ID: 6EWM) was retrieved from the protein data bank (PDB) as a PDB file. The crystal structure has a resolution of 1.4 Å. The protein is homodimer having 191 amino acid residues in total [13] Figure 1.
Figure 1: 3D structure of the heme-binding protein HmuY of P.gingivalis complexed with MES and sulfate ions (PDB ID: 6EWM).
For preparing the receptor, all the heteroatoms, water, and ions were removed to obtain the native receptor protein. In addition, polar hydrogens were added and the charges were assigned.
Ligands preparation
A series of 10 common essential oil molecules and 10 coumarin derivatives were downloaded from PubChem and Chemspider datasets as SDF files and then converted to PDB format using Open Babel software [14]. After, the downloaded ligands were energy-minimized. Table 1 enumerates the tested ligands along with their structure. The selection criteria of ligands were on the observation that essential oils demonstrated their efficacy against QS of certain bacteria [15-17]. On the other hand, coumarin inhibited the QS of P.gingivalis In silico and in vitro [12].
Table 1: List of the selected compounds to be docked against HmuY. The table lists the 2 groups (essential oils and coumarin derivatives) along with the corresponding structure.
Molecular docking
Protein and ligand preparation, energy minimization as well as molecular docking were performed using UCSF Chimera software (version 1.16) and its built-in autodock vina interface [18,19]. We performed a blind docking with a grid box covering all the receptor structures with dimensions 50 × 43 × 72 Å and centered at -2.355, 6.734 and 24.335 of X, Y and Z coordinates.
Data visualization
To explore the docking interaction of best confirmation of the ligands with the highest binding affinities of both essential oils and coumarin derivatives, PyMOL [20], (surface mode) and Proteinplus webserver (2D diagram) [21], were employed for visualizations.
ADMET prediction
In order to evaluate the pharmacokinetics as well as drug-likeness of the selected ligands, the ADMETsar server was utilized [22]. Moreover, the ProTox-II platform [23], was used to assess the toxicity profiles of only the best docking compound of each group.
Instead of the application of antibiotics that have no impact against pathogenic bacterial biofilms, some natural products emerge as potent factors interfering with QS and thus biofilm formation [24]. P.gingivalis is one bacterium found in oral biofilms that are correlated with many burden disorders like AD [25]. Therefore, upon targeting the QS pathway and preventing biofilm formation, a huge number of inflicted populations will benefit especially if the cure is of natural origin.
Docking analysis
The binding affinities of the selected ligands were tested using the AutoDock vina interface within UCSF Chimera software. As shown in Table 2, all of the tested ligands belonging to essential oils exhibited higher binding affinity values compared to the reference inhibitor MES whose value was -5.5 kcal/mole. Cardinal gave maximum binding affinity of all essential oil molecules (-8.1 kcal/mole). The rest compounds' values were between -6.2 kcal/mole to -6.7 kcal/mole.
| Table 2: Molecular docking results of all selected compounds from the two groups against the target protein 6EWM. | |||
| Ligand | Binding affinity | Ligand | Binding affinity |
| Camphor | -6.5 | Coumarin | -7 |
| Menthol | -6.5 | Derivative 1 | -10.4 |
| Fenchone | -6.4 | Derivative 2 | -8.5 |
| Terpineol | -6.7 | Derivative 3 | -8.5 |
| Anethole | -6.2 | Derivative 4 | -8.9 |
| Cardinol | -8.1 | Derivative 5 | -9.8 |
| R-Carvone | -6.4 | Derivative 6 | -8.7 |
| S-Carvone | -6.6 | Derivative 7 | -8.7 |
| Thymol | -6.6 | Derivative 8 | -9.8 |
| Trans-linalool oxide | -6.2 | Derivative 9 | -8.9 |
| MES | -5.5 | Derivative 10 | -9.2 |
Conversely, coumarin and its derivatives had higher values of binding affinity than essential oils and the reference inhibitor MES (ranged from -8.5 kcal/mole to -10.4 kcal/mole). It should be noted that coumarin derivatives are better than coumarin moiety which gave an inhibition value of -7 kcal/mole.
It seems reasonable for coumarins to have higher free energy values than essential oils as they have at least two rings in their structure which make them more suitable to be fitted in the active center of the protein.
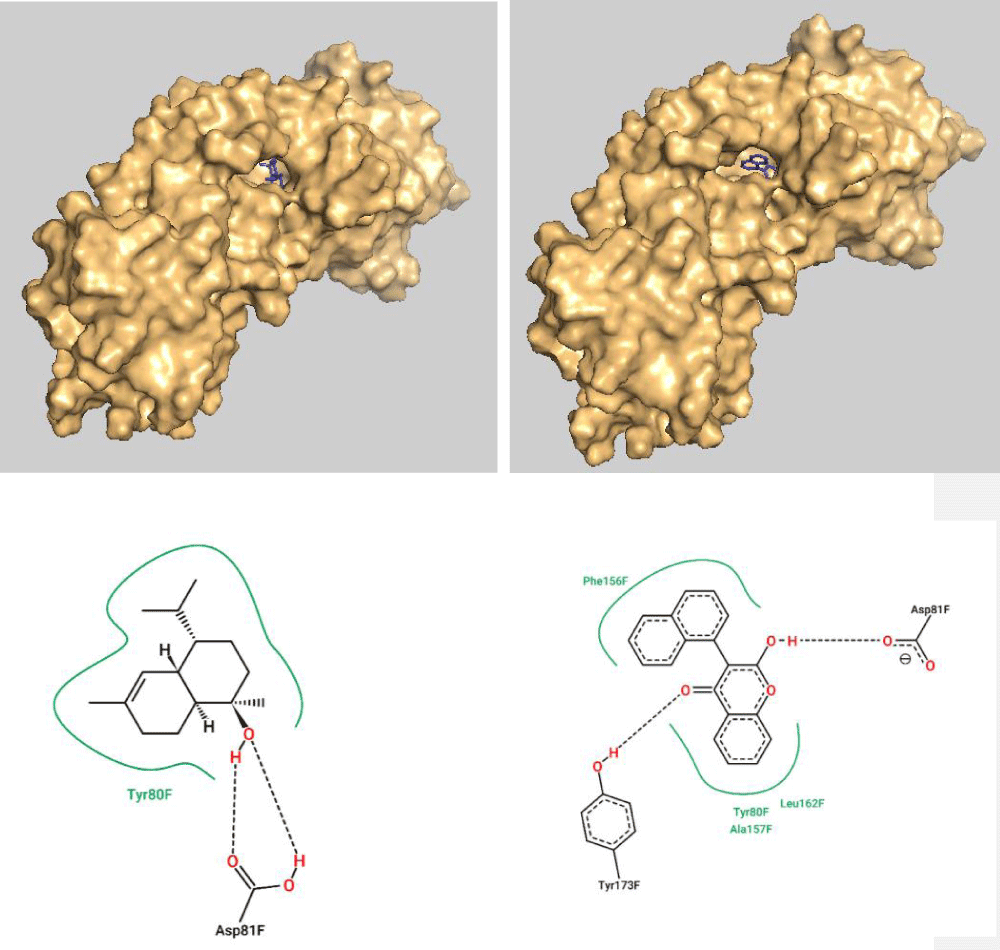
The top molecules with the highest binding energy of the two groups (cardinol and coumarin derivative 1) were analyzed further for their interaction with the active site of the target protein HmuY. Figure 2 illustrates the surface view of the receptor with the cardinol and coumarin derivative 1 bound to its active pocket as depicted using PyMOL software. In addition, the 2D diagram of interaction is also shown in Figure 2 as predicted via proteins plus server.
Since coumarin derivative 1 has more energy values in comparison with the essential oil cardinol, it has more H-bonds and VdW interactions within the active site of the target protein HmuY. Coumarin derivative 1 has more interactions with Tyr 173 (via H-bonds) and Leu 162, Phe 156, and Ala 157 (through VdW interactions) while Asp 81, as well as Tyr 80, appear to have mutual interactions with both of them (Figure 2).
Figure 2: Surface view of the target protein with the cardinol (Top left) and coumarin derivative (Top right) bound to the active pocket. Also, the 2D diagram of cardinol interaction (bottom left) and coumarin derivative 1 (bottom right) with the active site residues.
Prediction of ADMET parameters
It is necessary to predict and calculate the pharmacokinetic properties of the docked ligands to figure out their druggability. Table 3 lists the ADME properties of all ligands obtained from the ADMETsar 2.0 webserver.
As shown in Table 3, all of the tested ligands in the two groups are in a domain, i.e. they fit well to Lipinski's rule of five. This means the good druggability profile of the selected compounds. However, a toxicity profile must be carried out to evaluate the possible toxicity issues of the docked molecules to be lead candidates as inhibitors of the QS of P.gingivalis. Table 4 depicts the toxicity paramters of cardinol and coumarin derivative 1 (the best-docked molecules).
| Table 3: Obtained ADME properties of all selected ligands in the two groups along with their applicability domain. | ||||||
| Ligand | MW | AlogP | HA | HD | RB | Applicability Domain |
| Essential oils | ||||||
| Camphor | 152.24 | 2.4 | 1 | 0 | 0 | In domain |
| Menthol | 156.27 | 2.44 | 1 | 1 | 1 | In domain |
| Fenchone | 152.24 | 2.4 | 1 | 0 | 0 | In domain |
| Terpineol | 154.25 | 2.5 | 1 | 1 | 1 | In domain |
| Anethole | 148.2 | 2.73 | 1 | 0 | 2 | In domain |
| Cardinol | 222.37 | 3.78 | 1 | 1 | 1 | In domain |
| R-Carvone | 150.22 | 2.49 | 1 | 0 | 1 | In domain |
| S-Carvone | 150.22 | 2.49 | 1 | 0 | 1 | In domain |
| Thymol | 150.22 | 2.82 | 1 | 1 | 1 | In domain |
| Trans-linalool oxide | 170.25 | 1.88 | 2 | 1 | 2 | In domain |
| MES | 195.24 | -0.79 | 4 | 1 | 3 | In domain |
| Coumarin derivatives | ||||||
| Coumarin | 146.14 | 1.79 | 2 | 0 | 0 | In domain |
| Derivative 1 | 288.3 | 4.32 | 3 | 1 | 1 | In domain |
| Derivative 2 | 253.26 | 2.86 | 4 | 0 | 2 | In domain |
| Derivative 3 | 257.68 | 3.51 | 3 | 0 | 1 | In domain |
| Derivative 4 | 273.29 | 1.19 | 4 | 0 | 2 | In domain |
| Derivative 5 | 272.3 | 2 | 4 | 1 | 1 | In domain |
| Derivative 6 | 239.23 | 2.56 | 4 | 1 | 1 | In domain |
| Derivative 7 | 268.27 | 3.17 | 4 | 1 | 2 | In domain |
| Derivative 8 | 271.32 | 2.35 | 3 | 0 | 2 | In domain |
| Derivative 9 | 339.3 | 2.71 | 7 | 0 | 3 | In domain |
| Derivative 10 | 397.43 | 1.68 | 6 | 1 | 8 | In domain |
| MW: Molecular Weight; HA: Hydrogen Acceptor; HD: Hydrogen Donor; RB: Rotatable Bonds. | ||||||
| Table 4: ProTox-II server results of toxicity profiles of cardinol and coumarin derivative 1. | ||||
| Target | Cardinol | Coumarin derivative 1 | ||
| Prediction | Probability | Prediction | Probability | |
| Hepatotoxicity | Inactive | 0.82 | Inactive | 0.67 |
| Carcinogenicity | Inactive | 0.66 | Active | 0.55 |
| Immunotoxicity | Active | 0.69 | Inactive | 0.81 |
| Mutagenicity | Inactive | 0.91 | Inactive | 0.83 |
| Cytotoxicity | Inactive | 0.87 | Inactive | 0.62 |
| Aryl hydrocarbon Receptor (AhR) | Inactive | 0.98 | Active | 0.55 |
| Androgen Receptor (AR) | Inactive | 0.87 | Inactive | 0.98 |
| Aromatase | Inactive | 0.92 | Inactive | 0.7 |
| Estrogen Receptor Alpha (ER) | Inactive | 0.8 | Inactive | 0.53 |
| Estrogen Receptor Ligand Binding Domain (ER-LBD) | Inactive | 0.83 | Inactive | 0.74 |
| Peroxisome Proliferator-Activated Receptor Gamma (PPAR-Gamma) | Inactive | 1 | Inactive | 0.59 |
| Heat shock factor response element (HSE) | Inactive | 0.69 | Inactive | 0.97 |
| Mitochondrial Membrane Potential (MMP) | Active | 0.5 | Active | 0.6 |
| Phosphoprotein (Tumor Supressor) p53 | Inactive | 0.99 | Inactive | 0.75 |
Concerning toxicity assessment, cardinol exhibited slight immunogenicity (probability 0.69) and showed activity toward MMP. Similarly, coumarin derivative 1 was found to have carcinogenicity (probability 0.55) and activity toward AhR as well as MMP. In short, the predicted toxicity of cardinol is class IV whereas coumarin derivative 1 is class III as calculated by the ProTox-II platform. This work suggests the application of the mentioned ligands as a means to control the biofilm of P.gingivalis found within the oral cavity and its associated burden diseases such as dental plaques, Alzheimer's disease, and atherosclerosis. By approving their potency In silico, the best 2 ligands should find their way in vitro setting to confirm the computational prediction by wet lab experiments as coupled by [12] .
According to the data obtained in this study, we concluded the stable inhibition of P.gingivalis QS via some natural essential oil molecules (best of which was cardinol with a binding affinity of -8.1 kcal/mole) and some coumarin derivatives. The best of which was derivative 1 exhibiting a binding affinity of -10.4 kcal/mole. Furthermore, the docked ligands demonstrated good ADME properties which candidate them superiorly above the reference inhibitor MES and even the native coumarin. However, toxicity prediction results revealed that cardinol was immunogenic and coumarin derivative 1 was carcinogenic. Thereby, we recommend after confirmation of results in vitro experiments to assay the immunogenicity and carcinogenicity of the cardinol and coumarin derivative 1 as well or at least can be utilized as a lead pharmacophore through which safer pharmaceuticals can be designed and improved.
- Bowen WH, Burne RA, Wu H, Koo H. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018 Mar;26(3):229-242. doi: 10.1016/j.tim.2017.09.008. Epub 2017 Oct 30. PMID: 29097091; PMCID: PMC5834367.
- Rath S, Bal SCB, Dubey D. Oral Biofilm: Development Mechanism, Multidrug Resistance, and Their Effective Management with Novel Techniques. Rambam Maimonides Med J. 2021 Jan 19;12(1):e0004. doi: 10.5041/RMMJ.10428. PMID: 33478627; PMCID: PMC7835112.
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135-87. doi: 10.1111/j.1600-0757.2005.00107.x. PMID: 15853940.
- Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010 Jul;8(7):471-80. doi: 10.1038/nrmicro2381. PMID: 20514044.
- Kanagasingam S, Chukkapalli SS, Welbury R, Singhrao SK. Porphyromonas gingivalis is a Strong Risk Factor for Alzheimer's Disease. J Alzheimers Dis Rep. 2020 Dec 14;4(1):501-511. doi: 10.3233/ADR-200250. PMID: 33532698; PMCID: PMC7835991.
- Zhang J, Xie M, Huang X, Chen G, Yin Y, Lu X, Feng G, Yu R, Chen L. The Effects of porphyromonas gingivalis on Atherosclerosis-Related Cells. Front Immunol. 2021 Dec 23;12:766560. doi: 10.3389/fimmu.2021.766560. PMID: 35003080; PMCID: PMC8734595.
- Zhou Y, Luo GH. porphyromonas gingivalis and digestive system cancers. World J Clin Cases. 2019 Apr 6;7(7):819-829. doi: 10.12998/wjcc.v7.i7.819. PMID: 31024953; PMCID: PMC6473131.
- Jia L, Han N, Du J, Guo L, Luo Z, Liu Y. Pathogenesis of Important Virulence Factors of porphyromonas gingivalis via Toll-Like Receptors. Front Cell Infect Microbiol. 2019 Jul 18;9:262. doi: 10.3389/fcimb.2019.00262. PMID: 31380305; PMCID: PMC6657652.
- Berger D, Rakhamimova A, Pollack A, Loewy Z. Oral Biofilms: Development, Control, and Analysis. High Throughput. 2018 Aug 31;7(3):24. doi: 10.3390/ht7030024. PMID: 30200379; PMCID: PMC6163956.
- Li XH, Lee JH. Antibiofilm agents: A new perspective for antimicrobial strategy. J Microbiol. 2017 Oct;55(10):753-766. doi: 10.1007/s12275-017-7274-x. Epub 2017 Sep 28. PMID: 28956348.
- Lu L, Hu W, Tian Z, Yuan D, Yi G, Zhou Y, Cheng Q, Zhu J, Li M. Developing natural products as potential anti-biofilm agents. Chin Med. 2019 Mar 20;14:11. doi: 10.1186/s13020-019-0232-2. PMID: 30936939; PMCID: PMC6425673.
- He Z, Jiang W, Jiang Y, Dong J, Song Z, Xu J, Zhou W. Anti-biofilm activities of coumarin as quorum sensing inhibitor for Porphyromonas gingivalis. J Oral Microbiol. 2022 Mar 29;14(1):2055523. doi: 10.1080/20002297.2022.2055523. PMID: 35368854; PMCID: PMC8967191.
- Bielecki M, Antonyuk S, Strange RW, Smalley JW, Mackiewicz P, Śmiga M, Stępień P, Olczak M, Olczak T. Tannerella forsythia Tfo belongs to porphyromonas gingivalis HmuY-like family of proteins but differs in heme-binding properties. Biosci Rep. 2018 Oct 22;38(5):BSR20181325. doi: 10.1042/BSR20181325. PMID: 30266745; PMCID: PMC6200708.
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011 Oct 7;3:33. doi: 10.1186/1758-2946-3-33. PMID: 21982300; PMCID: PMC3198950.
- Hançer Aydemir D, Çifci G, Aviyente V, Boşgelmez-Tinaz G. Quorum-sensing inhibitor potential of trans-anethole aganist Pseudomonas aeruginosa. J Appl Microbiol. 2018 Sep;125(3):731-739. doi: 10.1111/jam.13892. Epub 2018 Jun 26. PMID: 29694695.
- Zhang Y, Kong J, Xie Y, Guo Y, Cheng Y, Qian H, et al. Essential oil components inhibit biofilm formation in Erwinia carotovora and Pseudomonas fluorescens via anti-quorum sensing activity. LWT 2018; 92: 133-139. https://doi.org/10.1016/j.lwt.2018.02.027.
- Szabó MA, Varga GZ, Hohmann J, Schelz Z, Szegedi E, Amaral L, Molnár J. Inhibition of quorum-sensing signals by essential oils. Phytother Res. 2010 May;24(5):782-6. doi: 10.1002/ptr.3010. PMID: 19827025.
- Butt SS, Badshah Y, Shabbir M, Rafiq M. Molecular docking using chimera and autodock vina software for nonbioinformaticians. JMIR Bioinformatics and Biotechnology. 2020; 1:e14232.
- Li Q, Shah S. Structure-based virtual screening. Protein Bioinformatics, Springer; 2017; 111-124.
- DeLano WL. Pymol: An open-source molecular graphics tool. CCP4 Newsl Protein Crystallogr 2002; 40: 82-92.
- Schöning-Stierand K, Diedrich K, Fährrolfes R, Flachsenberg F, Meyder A, Nittinger E, Steinegger R, Rarey M. Proteinsplus: interactive analysis of protein-ligand binding interfaces. Nucleic Acids Res. 2020 Jul 2;48(W1):W48-W53. doi: 10.1093/nar/gkaa235. PMID: 32297936; PMCID: PMC7319454.
- Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z, Li W, Liu G, Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019 Mar 15;35(6):1067-1069. doi: 10.1093/bioinformatics/bty707. PMID: 30165565.
- Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018 Jul 2;46(W1):W257-W263. doi: 10.1093/nar/gky318. PMID: 29718510; PMCID: PMC6031011.
- Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019 May 16;8:76. doi: 10.1186/s13756-019-0533-3. PMID: 31131107; PMCID: PMC6524306.
- Olsen I. porphyromonas gingivalis-Induced Neuroinflammation in Alzheimer's Disease. Front Neurosci. 2021 Oct 14;15:691016. doi: 10.3389/fnins.2021.691016. PMID: 34720846; PMCID: PMC8551391.