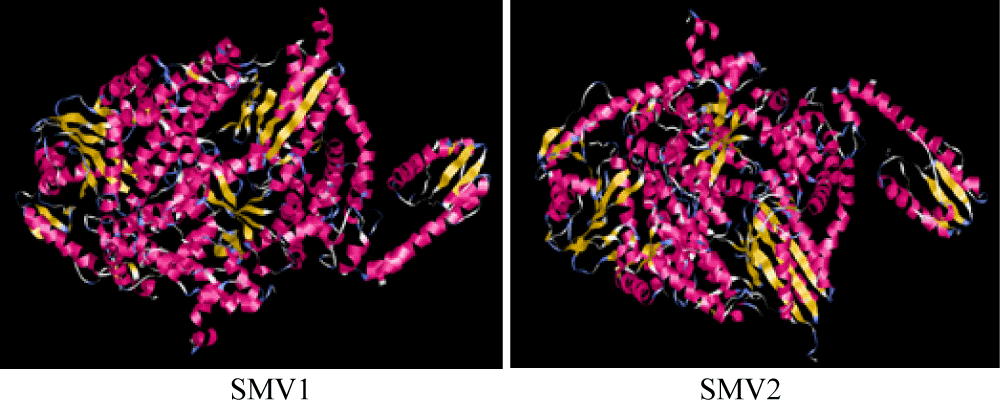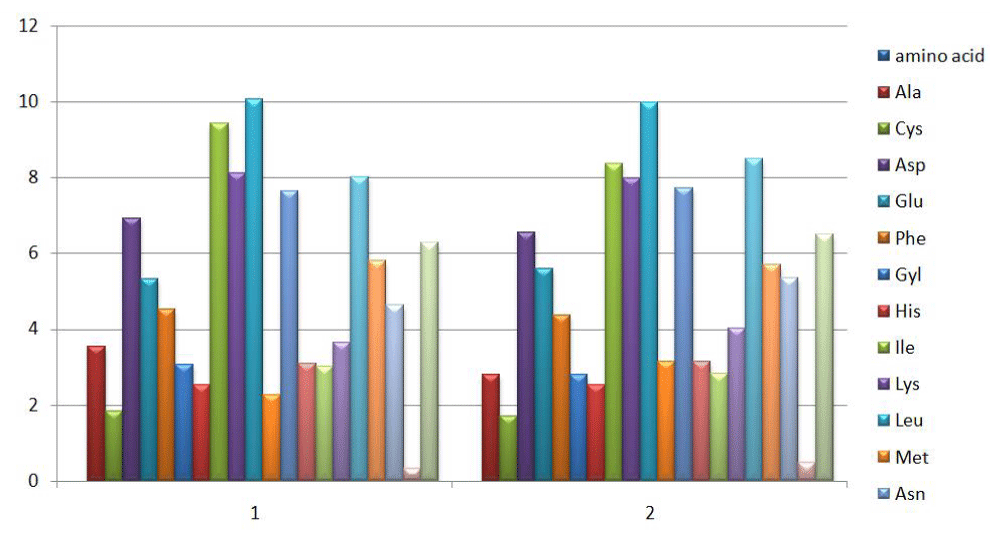More Information
Submitted: May 18, 2021 | Approved: June 12, 2021 | Published: June 14, 2021
How to cite this article: Singh N, Narula B, Ujinwal M, Langyan S. Pigeonpea sterility mosaic virus a green plague-Current status of available drug and new potential targets. Ann Proteom Bioinform. 2021; 5: 008-026.
DOI: 10.29328/journal.apb.1001013
Copyright License: © 2021 Singh N, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Drug targets; Emaravirus; Pigeonpea; Protein structure SMD; SMV1 and SMV2
Abbreviations: AFLP: Amplified Fragment Length Polymorphism; DAS-ELISA: Double Antibody Sandwich ELISA; DIBA: Dot Immunobinding Assay; ELISA: The Enzyme-Linked Immunosorbent Assay; RT-PCR: Reverse Transcription Polymerase Chain Reaction; SMD: Sterility Mosaic Disease; SMV: Sterility Mosaic Virus
Pigeonpea sterility mosaic virus a green plague-Current status of available drug and new potential targets
Nisha Singh1*, Bhawna Narula1, Megha Ujinwal1 and Sapna Langyan2
1ICAR, National Institute for Plant Biotechnology, New Delhi-110012, India
2ICAR, National Bureau of Plant Genetic Resources, New Delhi-110012, India
*Address for Correspondence: Nisha Singh, ICAR, National Institute for Plant Biotechnology, New Delhi-110012, India, Email: [email protected]
Pigeonpea is one of the important legume crops with high protein content and nutritional traits. It has enormous potency for its widespread adoption by farming communities. It is affected by various kinds of biotic and abiotic stresses. In the context, of biotic stresses Sterility mosaic disease (SMD) is one of the severe diseases in pigeonpea which ultimately lead to the drastic yield loss. The virus belongs to the genus Emaravirus, family- Fimoviridae. SMD is associated with two diverse types of Emaravirus, Pigeonpea sterility mosaic virus1 (PPSMV-1) and Pigeonpea sterility mosaic virus 2 (PPSMV-2). It is transmitted by the mite (Aceria cajani), mainly environmental contributing to the feasibility for the mites for the inoculation of the virus. The SMD is mainly governed by two genes SV1 that includes the dominant allele and serves as an inhibitory action on the resistance of the SV2. Methods for identification of the virus include RT-PCR, DIBA and ELISA using alkaline phosphatase or penicillinase. To control SMV disease farmers generally adopted intercropping methods. There are few potential drugs have been identified for the administration of the disease such as 0.1% Fenazaquin, Dicofol, Imidacloripid, Carbosulfan; Spiromesifin includes the inhibition of the mite inoculation on the pigeonpea plant. The present review describes compressive and systematic insights on SMV protein targets and potential drugs that could be utilized as the presumed drug targets for the finding of true drugs against the SMD in pigeonpea.
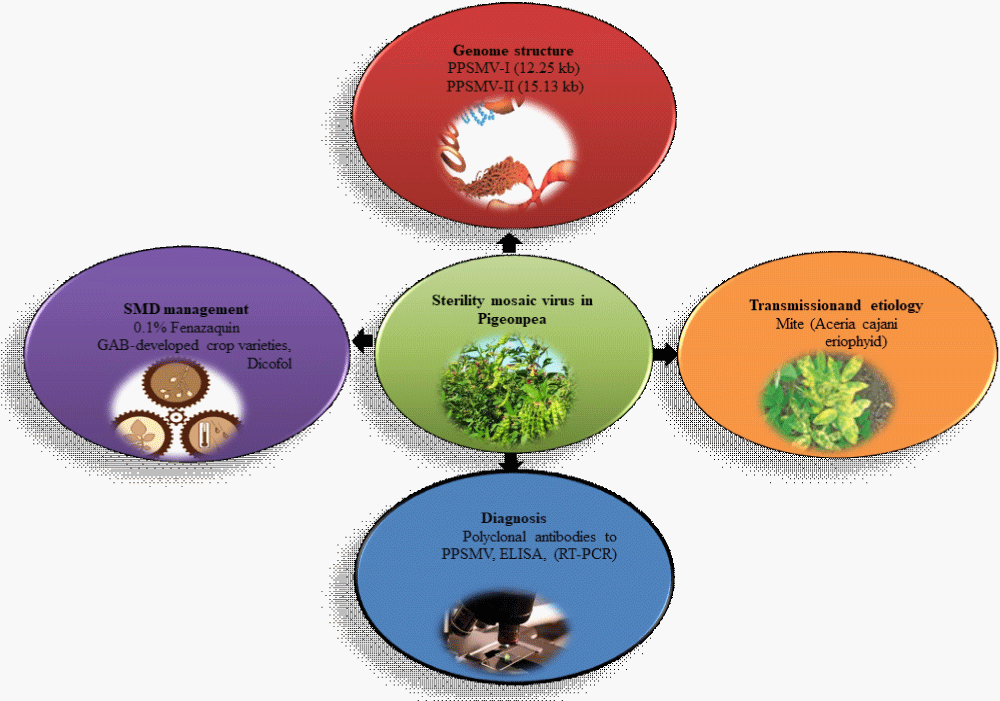
Pigeonpea (Cajanus Cajan L. Millsp.), also known as tur or red gram, a perennial shrub and tropical legume which enriches the soil through symbiotic nitrogen fixation [1]. In India, it is one of the important crops after chickpea and predominates the area of ~4.89 million tons (mt) and productivity is 762.4 kg ha–1 respectively [2]. It is a cross pollinated and diploid legume with 22 chromosomes and 858 Mb, encodes a total of 47,180 protein-coding genes [3]. It is mainly known for its high protein content (vitamin B, carotene and ascorbic acid), with a substantial amount of amino acid content up to 43.61% [4]. Despite being the most populated crop, it faces several environmental challenges (abiotic and biotic stress). In abiotic stress (low and high moisture, salinity and water-logging hypoxia) and biotic stresses (fusarium wilt, sterility mosaic and several insect pests including pod borer insects, maruca, stresses) which affects the yield of the plant [5]. Many diseases have led to low yield in the pigeonpea such as root-knot infestations (Meloidogyne spp.); Wilt (Fusarium udum) [6]. Sterility Mosaic Diseases are one of the biotic stresses which are highly disastrous and endemic in almost all agro-ecologies of pigeonpea growing in India. Apart from this, reniform nematodes (Rotylenchulus.spp.) also increases susceptibility to fusarium wilt and Sterility Mosaic Disease (SMD). The SMD is a major restriction on the production of pigeonpea and other legumes. SMD was first described in 1931 in Pusa, Bihar State and is mostly regional to India, Nepal, Bangladesh and Myanmar [7]. If infection of SMD occurs at an early stage i.e., less than 45-day-old plants, it leads to loss 95% - 100% yield [8]. SMD is one of repression on the pigeonpea that leads to the yield loss in consistency, thus leading to the epidemics. The symptomatic features of the seed are the stunting, bushy and pale green appearance of the plant with mosaic, mottling and reduced size of the leaves, but severely affected plants are seen with complete sterility of pods [9]. Pigeonpea and a few wild species of Cajanus were found to support the vector Aceria cajani(Acari: Arthropoda) , the eriophyid mite. Many studies conducted in the last few decades showed that in nature, A. cajani populations were almost exclusively observed on SMD-infected pigeonpea, but not on healthy plants, indicating a strong communalistic relationship between the virus-infected plants and the vector. The epidemiology of SMD involves the virus, mite vector, cultivar and environmental conditions. Infected perennial and volunteer plants serve as a source for both the virus and its vector mites and play an important role in the disease cycle. Wild Cajanus serves useful genes which have the greater potential for crop improvement. Several tools have been developed using the genomics approach to boost the genetic performance such as different types of molecular markers were added to trace the disease resistance varieties [10]. Many genetic improvement techniques are evolved i.e., pure line selection from germplasm, hybridization followed by pedigree selection and mutation breeding. Management of the disease with very rare drugs has been found to be effective. The biological control can be done through use of entomopathogenic fungi [11]. In this review we provide an outline of SMV including its symptoms, transmission, etiology, genome organization, and proteomic structure with a list of few available and potent drugs for the management and control of SMV in pigeonpea crop improvement programs (Figure 1).
Figure 1: Overview of sterility mosaic virus, genome organization, transmission and etiology, diagnosis and disease management.
The sterility mosaic disease in pigeonpea
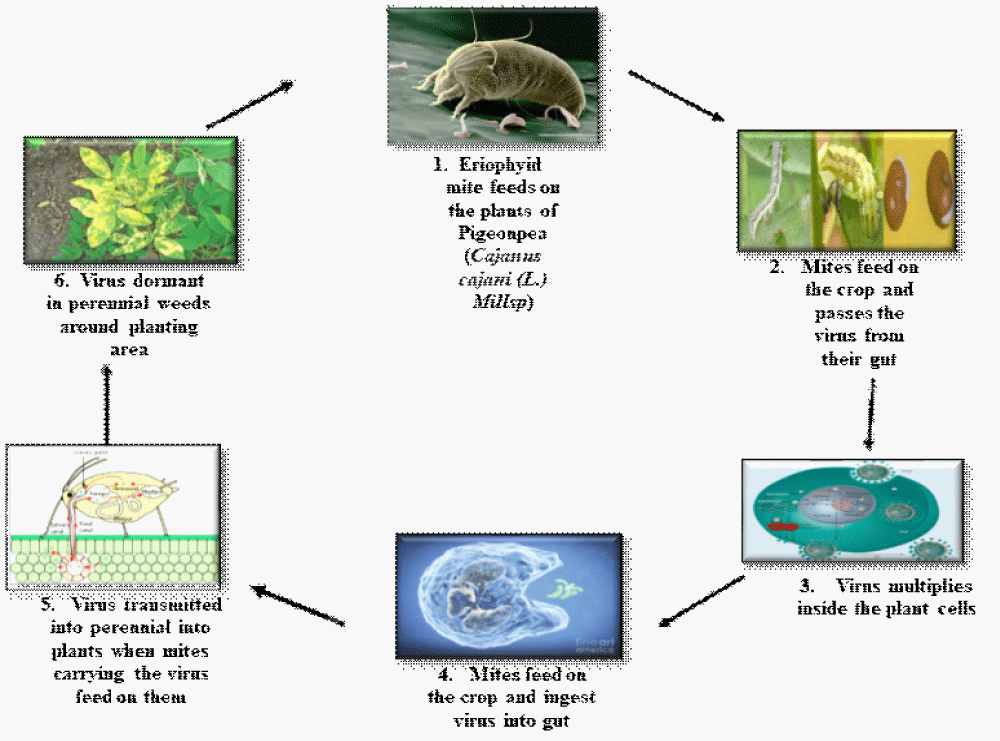
SMD is one of the major diseases in the Indian subcontinent associated with various factors that lead to different infections and it is transmitted by A. cajani. The main susceptible parts like branches show the mottling of the disease and diverse degree of mosaic affecting sterility of the infected plants [13]. Sterility Mosaic Disease is considered as one the major biotic stress causing disastrous damage to the yield leading to loss up to 95% [14]. A contingent agent of the disease is Emaravirus. The disease is also designated as 'green plague' since infected plants remain in the habitat [15]. SMD is characterized by one or more features such as partial cessation of production of flowers (sterility), mosaic or chlorotic ringspot. Symptoms of the leaves include excessive vegetative growth, stunting and decreased leaf size. Symptoms often depend on the infection time. Infection of susceptible species at the early stage of crop development, genotypes (< 30- to 45-day-old plants) result in characteristic disease expression symptoms within 10 to 15 days and nearly every other day complete cessation of the flowering process, but leaves plants grow, symptoms become masked [16]. The population of mites, their life cycle and the incidence of disease were observed to be affected by seasonal temperature fluctuations; relative humidity, direction of wind, speed and rainfall etc. [17]. SMV enters the plant cells through the mites which ejaculate the virus into the natural openings of the plant surface resulting in mechanical injury. Upon SMV entry, the viral genomic RNA is released and translated. Following translation, the viral proteins, and particles assemble new virus progeny and move to adjacent cells. Aphids spread the virus, relatively in a short time, after feeding on a virus-infected plant; mites instantly transfer to a healthy plant and transmit (spread) the virus as they feed on the other plant (Figure 2). Seeds are the source of primary inoculums, while secondary spreads are by aphids which happen at a rapid pace. Seed transmission efficiency varies by cultivar, with the incidence of seed transmission being higher in plants infected before flowering [18]. 5% of the transmission occurs in the majority of commercial cultivars produced, with older cultivars varying from no transmission to 75% transmission [19].
Figure 2: Depicting the transmission of virus by eriophyid mite which inoculates the pigeonpea plant passes the virus from their gut and feeds on another plant ultimately leading to the outspread of the disease.
Etiology and virus transmission
SMD has a distinctive attribute in stunted and bushy plants, reduced-size leaves with chlorotic rings or mosaic symptoms, and partial or complete cessation of flower production [20]. The causative agent of the disease is PPSMV, a single-stranded RNA genome virus with a segmented, negative sense, transmitted semi-persistently by the eriophyid mite Aceria cajani. Several genotypes of cultivated and wild relatives of pigeonpea are affected by PPSMV. The experimental hosts are N. Benthamian, N. Clevelandii, P. vulgaris, Chrozophorarottleri was used for the identification and spread of disease [21].
The virus and the vector are both intensely pigeonpea-specific and its few wild relatives are C.Scarabaeoides and C. cajanifolius. The presence of several PPSMV strains was suspected based on differential host reactions in different geographical areas. It spreads through the fields in an outbreak form under favorable conditions. Moreover, infection predisposes the plants to subsequent infection by fungal diseases and spider mite colonization [22]. The prevalence of asymptomatic field infections is normal and often much higher in areas of incidence of illness dependent on apparent foliar symptoms [23]. Differential host reactions are based on different regional reactions. The presence of different types of PPSMV strains can be detected in areas where pigeonpea is cultivated after a long gap. The epidemiology of SMD involves the virus, vector mite, cultivar and environmental conditions. Perennial infected and volunteer plants both serve as a source for the virus. Its vector mites have an important role to play in the disease cycle [24]. A. cajani was up to 53% but was 100% when > 5 mites were used per plant [25]. A. cajani has a short length of 2.03 um (approx.) which enables transmission to the epidermal and the underneath mesophyll cells [26]. The multiplication of mites on SMD-affected plants was very high compared to healthy pigeonpea plants. PPSMV has an incubation time of eight days for the expression of pigeonpea symptoms [27]. A number of central research foci, captivating research to determine critical pathogen targets for control, novel methodologies and methods of delivery, are evolving that will provide a strong basis for disease management into the future. As described below, assertive research and implementation for the development of natural resistance has lead to development of resistance varieties of pigeonpea from various national and international crop breeding programs.
Detection of SMD in pigeonpea plant
SMD symptoms are particular and show a diagnostic trait for its identification. Several attempts have been made since various strategies have been done for the detection of the disease agent to be characterized. The diagnosis of SMD and the selection of germplasm resistant to SMD are based on symptom expression. However, this diagnosis is complicated by the fact that many abiotic and biotic variables are governed in symptom expression and that pigeonpea is cross-pollinated so that genotypic heterogeneity, caused by cross-pollination, also plays an important role in symptomatology [28].
The host polyphenolic compounds on the pod surface which interfere with the stability and infectivity of the SMD agent, which obstructs virus infectivity and purification of the virus particle. The purification procedure for minimizing the effects of components by extraction of infected leaves in buffers containing chelating and reducing agents, high concentrations of nonionic detergent, and the precipitation. Purification was achieved by quasi-equilibrium zoning centrifugation Sucrose and CsCl gradients [29]. In order to detect the presence of PPSMV from their virulent vector using whole mite extract as an antigen, serological diagnostic techniques such as DAS-ELISA (double antibody sandwich ELISA) and DIBA (dot immunobinding assay) were developed. For virus detection in vector mites, both DAS-ELISA and DIBA was susceptible [30]. Polyclonal antibodies to PPSMV are VLP (Virus like particles) preparations produced in rabbits have been very effective in the detection of PPSMV in plant tissues by double antibody sandwich (DAS) and direct antigen coating forms of ELISA using enzyme-labeled (alkaline phosphatase or penicillinase) immuno-gammaglobulin. At the genome level, using reverse transcription–polymerase chain reaction (RT-PCR) which uses primers based on sequence [31]. Recently, different approaches have been developed in the genomic regions responsible for genetic study and breeding traits using the Axiom 62K SNP array which allows the marker assisted breeding to transfer this trait into elite cultivars and germplasm [32]. The recent advances of omics technologies deliver large-scale multidimensional data sets, better to integrate and utilize them for the development of disease resistance pigeonpea cultivars [3].
Management of SMD in pigeonpea plant
The numerous chemical methods such as organophosphorus (Sulphur with Propargite combination, Salicylic acid and acaricides viz; 0.1% Fenazaquin) were used for control of disease but these were not economically beneficial because the crop is grown in marginal conditions. Various conventional breeding programmes have been conducted in South India which among 115 wild Cajanus accessions belonging to six species, C. albicans, C. platycarpus, C. cajanifolius, C. lineatus, C. scarabaeoides, and C. sericeus which revealed about resistance of accessions, ICP 15614, 15615, 15626, 15684, 15688, 15700, 15701, 15725, 15734, 15736, 15737, 15740, 15924, 15925, and 15926 and can be cross compatible with pigeonpea [33]. The existence of separate PPSMV strains/isolates in various locations, however, makes it difficult for the identification of the isolates. Genotypes that are SMD-resistant have an epidermal cell wall and thicker leaf cuticles than those of genotypes prone to mite’s broad-spectrum resistance [34]. PPSMV exhibits that recessive gene are the key cause of resistance. Resistant trait was governed by two independent non-allelic genes, SV1 and SV2 in which one-gene SV1 which is a dominant allele has an inhibitory effect on the character (resistance) that regulates the other (SV2) gene. Based on this, the presence in one locus of the dominant SV1 gene allele suppresses the action of the dominant SV2 (resistance) gene allele found at another locus, resulting in susceptible phenotypes [35].
New sources of resistance to Fusarium wilt and SMD have been identified in a mini-core collection of pigeonpea germplasm. Molecular markers based on AFLP (Amplified fragment length polymorphism) is a PCR-based technique that uses selective amplification of a subset of digested DNA fragments to generate and compare unique fingerprints for genomes of interest. Various QTLs for resistance to isolates have been analyzed using composite interval mapping, which shows the involvement of different genes that grant resistance to these isolates [36].
Genome organization of SMV
PPSMV is the seventh species in the genus of Emaravirus, family Fimoviridae, order Bunyavirales). It shows features which are associated with the Tospovirus members of the genera (Family: Bunyaviridae) and Tenuivirus, both consist of single-stranded RNA viruses encode proteins [37].PPSMVs shows resemblance in context to genomic organization and conserved 5′ and 3′-end sequences with other Emaraviruses. The phylogenetic analysis of the RNA-3 sequence of disease causing two viral agents such as PPSMV-I and PPSMV-II exhibited considerable variability among seven different geographical strains depicting four states in southern India [38]. PPSMV-1 was the first to be identified and, subsequently, another separate PPSMV-2 Emaravirus was also reported to be involved in SMD. Both PPSMV-1and PPSMV-2 consist of six genomic segments of RNA, namely RNA1 (7022 nucleotide in length) coding for RNA-dependent RNA polymerase (RdRp, 2295 amino acids); RNA2 (2223 nucleotide) coding for glycoprotein (GP, 649 amino acids); RNA3 (1442 nucleotide) coding for nucleocapsid protein (NP, 309 amino acids); RNA4 (1563 nucleotide) coding for p4 (MP, 362 amino acids); RNA5 (1689 nucleotide) coding for p4 movement protein (MP, 362 amino acids); (238 amino acids)[39].The genome structure is spherical and segmented shape. It consists of four linear segments with negative-sense and single-stranded RNA. The genome size is around 12.2 kb. The genome structure consists of RNA1 (7.0kb) and RNA2 (2.3 kb). The viral RNA dependent RNA polymerase (L) binds to a promoter on each encapsulated segment and transcribes the mRNAs and is capped by L protein during synthesis using cap snatching [40]. The protein sequences of SMV were retrieved from UniProt database (http://www.uniprot.org/). In order to understand the structural properties of SMV, MODELLER is used for homology or comparative modeling of protein three-dimensional structures. The alignment of SMV sequences to be modeled with known related structures is provided and MODELLER automatically calculates a model containing all non-hydrogen atoms. MODELLER implements comparative protein structure modeling by satisfaction of spatial restraints [41]. The protein structure involves the two type i.e.Emaravirus1 and Emaravirus2 of base pair 2294 in length. By computer assisted analysis using MODELLER. The 3D structure was developed by the interaction of amino acid residues in which SMV1 and SMV2 both showed an alpha helix of 16%. In SMV1 beta strand is one-fold higher than SMV2beta strand (10%). Total 2% of TM helix present in SMV2while absent in SMV1 (Figure 3).
Proteomic structure of SMV
Figure 3: Depicts the 3D structure of SMV1 structure (left) and SMV2 structure (right).
Analysis of primary structure and secondary structure
The amino acid composition of SMV1 and SMV2 were analyzed. The amino acid composition reveals that SMV1 has 2294 amino acids and molecular weight of 266245.80 Daltons whereas SMV2 has 2294 amino acids and molecular weight of 267897.20 Daltons (Figure 4).
Figure 4: Depicts the molecular weight of number of amino acid included in sequence of SMV1 (1) and SMV2 (2) in pigeonpea.
Secondary structure
a) Hydrophobicity
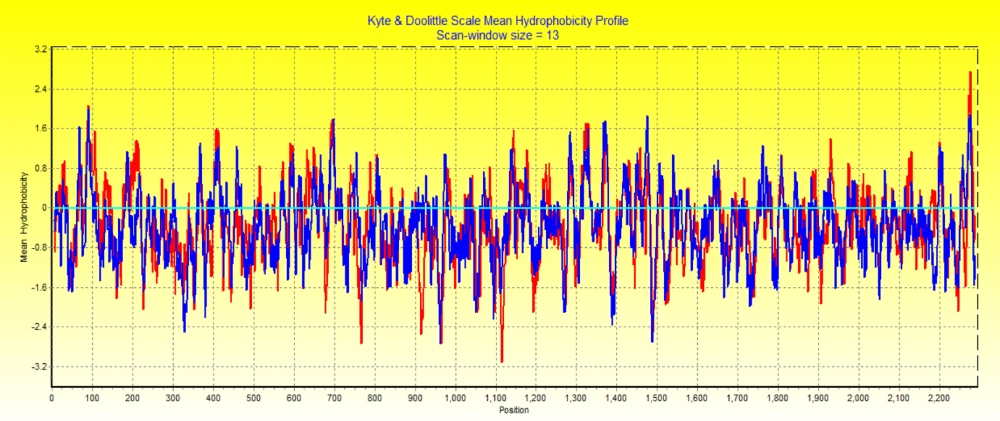
Kyte and Doolittle method is used to evaluate mean hydrophobicity as it can recognize surface exposed regions and transmembrane regions with interaction of protein region towards lipid bilayer. Lipid bilayer acts as a barrier around the cell, thus enable the virus to bind the host cells. Hydrophobic moment was determined for each residue by keeping the same window size for all of the amino acids. In this X-axis show position of each amino acid in sequence and Y-axis shows mean hydrophobicity. Value above zero is observed to have more amino acid at a particular position and give its mean hydrophobicity value (Figure 5).
Figure 5: Kyte-Doolitte hydrophobicity of two distinct Emaravirus SMV1 with the red region and SMV2withthe blue region.
b) Hydrophillicity
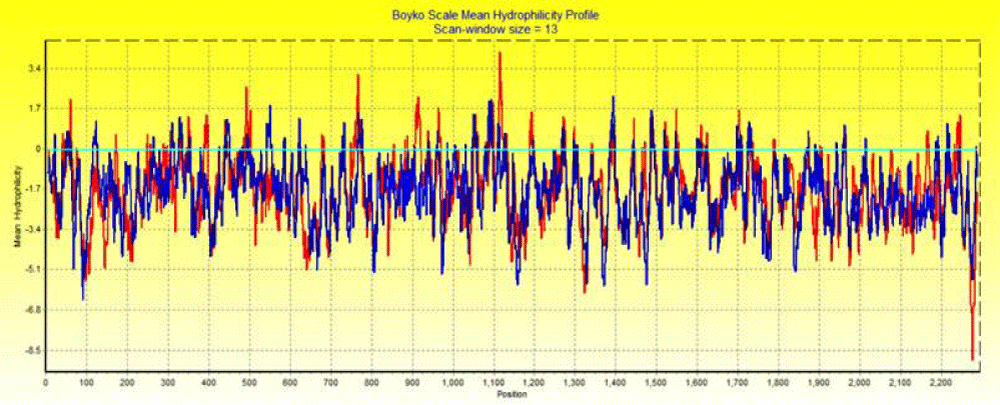
Boyko plot is a quantitative analysis of hydrophillicity of amino acids in SMV1 and SMV2. It measures the degree of interaction of the polar solvent with amino acids. Hydrophillicity indicates the quantitative analysis of amino acids of protein and helps of characterize domains of SMV1 and SMV2. Higher hydrophillicity indicates that residues are in contact with solvent and likely to occupy the outer surface of protein (Figure 6).
Figure 6: Bokyo plot hydrophillicity of two distinct Emaravirus SMV1 with red region and SMV2 with blue region.
New potential drug targets
SMV is one of the tropical diseases which are being treated with the non-SMD drugs however; the exact antimicrobial activity of the drug is not clearly understood. Search of new potential drugs only targets biochemical and metabolic pathways necessary for the mite survival [42]. Narrow and Broad-spectrum drugs are:
Narrow spectrum drugs only inhibit the multiplication of the unwanted pathogen by infecting the other virus by various strategies such as prohibiting protein, nucleic acid synthesis and metabolic pathways (Table 1).
| Table1: Summary on the narrow spectrum drugs for treatment of SMV in pigeonpea. | ||||
| S.No. | Drug name | Chemical structure | IUPAC Name: | CID number |
| 1. | Fenazaquin | 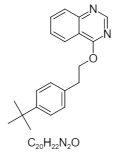 |
4-[2-(4-tert-butylphenyl)ethoxy]quinazoline | 86356 |
| 2. | Dicofol | 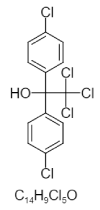 |
2,2,2-trichloro-1,1-bis(4-chlorophenyl)ethanol | 8268 |
| 3. | Spiromesifin | 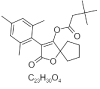 |
[2-oxo-3-(2,4,6-trimethylphenyl)-1-oxaspiro[4.4]non-3-en-4-yl] 3,3-dimethylbutanoate | 9907412 |
| 4. | Imidacloprid | 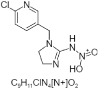 > > |
[[1-[(6-chloropyridin-3-yl)methyl]-4,5-dihydroimidazol-2-yl]amino]-hydroxy-oxoazanium | 101618973 |
| 5. | Carbosulfan | 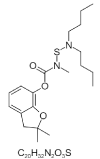 > > |
(2,2-dimethyl-3H-1-benzofuran-7-yl) N-(dibutylamino)sulfanyl-N-methylcarbamate | 41384 |
| 6. | Kelthane | 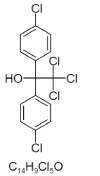 > > |
2,2,2-trichloro-1,1-bis(4-chlorophenyl)ethanol | 8268 |
| 7. | CEMBL251502 | 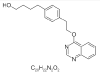 |
4-[4-(2-quinazolin-4-yloxy ethyl)phenyl]butan-1-ol | 44441989 |
| 8. | Chlorfenethol | 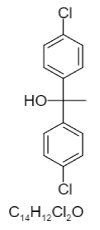 |
1,1-bis(4-chlorophenyl)ethanol | 6624 |
| 9. | Proclonol | 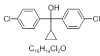 |
bis(4-chlorophenyl)-cyclopropylmethanol | 26450 |
| 10. | Brl 15268 | 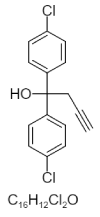 |
1,1-bis(4-chlorophenyl)but-3-yn-1-ol | 173320 |
| 11. | 2,2-Dichloro-1,1-bis(4-Chlorophenyl)Ethanol | 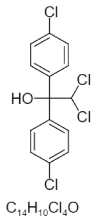 |
2,2-dichloro-1,1-bis(4-chlorophenyl)ethanol | 160701 |
| 12. | CHEMBL3186301 | 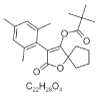 |
[2-oxo-3-(2,4,6-trimethylphenyl)-1-oxaspiro[4.4]non-3-en-4-yl] 2,2-dimethylpropanoate | 11348842 |
| 13. | Escitalopram | 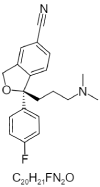 |
(1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-3H-2-benzofuran-5-carbonitrile | 2771 |
| 14. | Nitenpyram | 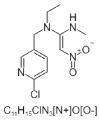 |
(NE)-N-[1-[(6-chloropyridin-3-yl)methyl]imidazolidin-2-ylidene]nitramide | 86287518 |
| 15. | Furathiocarb | 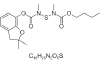 |
(2,2-dimethyl-3H-1-benzofuran-7-yl)N-[butoxycarbonyl(methyl)amino]sulfanyl-N-methylcarbamate | 47759 |
| 16. | Benfuracarb | 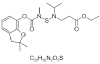 |
ethyl 3-[[(2,2-dimethyl-3H-1-benzofuran-7-yl)oxycarbonyl-methylamino]sulfanyl-propan-2-ylamino]propanoate | 54886 |
| 17. | Benzofuran | 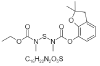 |
ethyl N-[(2,2-dimethyl-3H-1-benzofuran-7-yl)oxycarbonyl-methylamino]sulfanyl-N-methylcarbamate | 47756 |
| 18. | Carbamic acid | 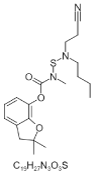 |
(2,2-dimethyl-3H-1-benzofuran-7-yl)N-[butyl(2-cyanoethyl)amino]sulfanyl-N-methylcarbamate | 3067994 |
Broad spectrum drugs targets and inhibits replication and development of the viral proteins such as polymerase and proteases by interfering with their cellular machinery (Table 2).
| Table 2: Summary on broad spectrum drugs for the treatment of SMV in pigeonpea. | ||||
| S.No | Drug name | Chemical structure | IUPAC Name: | Reference |
| 1. | Lactoferrin | 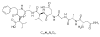 |
(2S)-2-[[(2S)-2-[[(2S)-2-[[(2S)-2-[[(2S)-3-carboxy-2-[[2-[[(2S)-2-[[(2S)-2,4-diamino-4-oxobutanoyl]amino]propanoyl]amino]acetyl]amino]propanoyl]amino]-3-methylbutanoyl]amino]propanoyl]amino]-3-phenylpropanoyl]amino]-3-methylbutanoic acid | 131676698 |
| 2. | Oleandomycin | 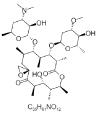 |
(3R,5R,6S,7S,8S,9S,12S,13R,14R,15R)-6-[(2S,3R,4S,6S)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy-14-hydroxy-8-[(2R,4S,5S,6S)-5-hydroxy-4-methoxy-6-methyloxan-2-yl]oxy-5,7,9,12,13,15-hexamethyl-1,11-dioxaspiro[2.13]hexadecane-10,16-dione | 72493 |
| 3. | Ampicillin | 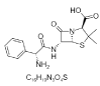 |
(2S,5R,6R)-6-[[(2R)-2-amino-2-phenylacetyl]amino]-3,3-dimethyl-7-oxo-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid | 6249 |
| 4. | Penicillin G | 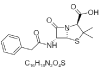 |
(2S,5R,6R)-3,3-dimethyl-7-oxo-6-[(2-phenylacetyl)amino]-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid | 5904 |
| 5. | Roxithromycin | 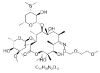 |
(3R,4S,5S,6R,7R,9R,10Z,11S,12R,13S,14R)-6-[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyl hexan-2-yl]oxy-14-ethyl-7,12,13-trihydroxy-4-[(2R,4R,5S,6S)-5-hydroxy-4-methoxy-4,6-dimethyl oxan-2-yl]oxy-10-(2-methoxyethoxymethyl imino)-3,5,7,9,11,13-hexamethyl-oxacyclotetradecan-2-one | 6915744 |
| 6. | Azithromycin | 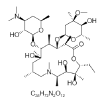 |
(2R,3S,4R,5R,8R,10R,11R,12S,13S,14R)-11-[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy-2-ethyl-3,4,10-trihydroxy-13-[(2R,4R,5S,6S)-5-hydroxy-4-methoxy-4,6-dimethyloxan-2-yl]oxy-3,5,6,8,10,12,14-heptamethyl-1-oxa-6-azacyclopentadecan-15-one | 447043 |
| 7. | Isoprenaline | 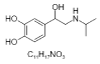 |
4-[1-hydroxy-2-(propan-2-ylamino)ethyl]benzene-1,2-diol | 3779 |
| 8. | Riboflavin | 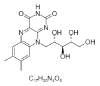 |
7,8-dimethyl-10-[(2S,3S,4R)-2,3,4,5-tetrahydroxypentyl]benzo[g]pteridine-2,4-dione | 493570 |
| 9. | Atropine | 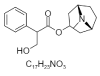 |
[(1S,5R)-8-methyl-8-azabicyclo[3.2.1]octan-3-yl]3-hydroxy-2-phenylpropanoate | 174174 |
| 10. | Albendazole | 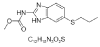 |
MethylN-(6-propylsulfanyl-1H-benzimidazol-2-yl)carbamate | 2082 |
| 11. | Neomycin B | 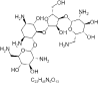 |
(2R,3S,4R,5R,6R)-5-amino-2-(aminomethyl)-6-[(1R,2R,3S,4R,6S)-4,6-diamino-2-[(2S,3R,4S,5R)-4-[(2R,3R,4R,5S,6S)-3-amino-6-(aminomethyl)-4,5-dihydroxyoxan-2-yl]oxy-3-hydroxy-5-(hydroxymethyl)oxolan-2-yl]oxy-3-hydroxycyclohexyl]oxyoxane-3,4-diol | 8378 |
| 12. | L-Tyrosylglycyl L-isoleucyl L- phenylalaniamide | 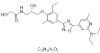 |
N-[(2S)-3-[4-[5-[2-(diethylamino)-6-methylpyridin-4-yl]-1,2,4-oxadiazol-3-yl]-2-ethyl-6-methylphenoxy]-2-hydroxypropyl]-2-hydroxyacetamide | 25074253 |
| 13. | CHEMBL330666; BDBM50014232; 6-(4-Ethynyl-Phenyl)-1,2,3,5,6,10b-Hexahydro-Pyrrolo[2,1-A]Isoquinoline | 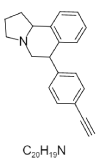 |
(6S,10bR)-6-(4-ethynylphenyl)-1,2,3,5,6,10b-hexahydropyrrolo[2,1-a]isoquinoline | 13903191 |
| 14. | Diazepine | 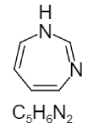 |
1H-diazepine | 166734 |
| 15. | Ningnanmycin | 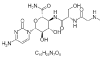 |
(2S,3S,4S,5R,6R)-6-(4-amino-2-oxopyrimidin-1-yl)-4,5-dihydroxy-3-[[(2S)-3-hydroxy-2-[[2-(methylamino)acetyl]amino]propanoyl]amino]oxane-2-carboxamide | 44588235 |
| 16. | Ribavirin | 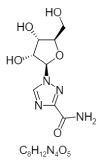 |
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,4-triazole-3-carboxamide | 37542 |
| 17. | Tylophorine | 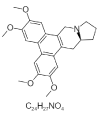 |
(13aS)-2,3,6,7-tetramethoxy-9,11,12,13,13a,14-hexahydrophenanthro[9,10-f]indolizine | 5090648 |
| 18. | Cryptopleurine | 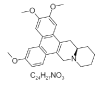 |
(14aR)-2,3,6-trimethoxy-11,12,13,14,14a,15-hexahydro-9H-phenanthro[9,10-b]quinolizine | 92765 |
| 19. | Antofine | 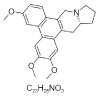 |
(13aR)-2,3,6-trimethoxy-9,11,12,13,13a,14-hexahydrophenanthro[9,10-f]indolizine | 639288 |
| 20. | Ribavirin monophosphate | 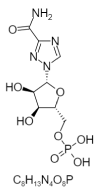 |
[(2R,3S,4R,5R)-5-(3-carbamoyl-1,2,4-triazol-1-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen phosphate | 100252 |
| 21. | Taribavirin | 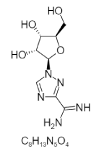 |
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,4-triazole-3-carboximidamide | 451448 |
| 22. | Leovirin | 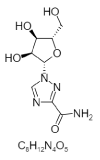 |
1-[(2S,3S,4R,5S)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,4-triazole-3-carboxamide | 460516 |
| 23. | Tylophorincine | 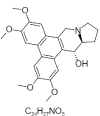 |
(13aS,14R)-2,3,6,7-tetramethoxy-9,11,12,13,13a,14-hexahydrophenanthro[9,10-f]indolizin-14-ol | 44443369 |
| 24. | Tobramycin | 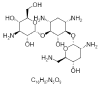 |
(2S,3R,4S,5S,6R)-4-amino-2-[(1S,2S,3R,4S,6R)-4,6-diamino-3-[(2R,3R,5S,6R)-3-amino-6-(aminomethyl)-5-hydroxyoxan-2-yl]oxy-2-hydroxycyclohexyl]oxy-6-(hydroxymethyl)oxane-3,5-diol | 36294 |
| 25. | TabavirinHcl | 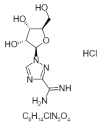 |
1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,2,4-triazole-3-carboximidamide;hydrochloride | 451447 |
| 26. | Dd-Rabvarin | 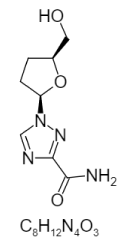 |
1-[(2R,5S)-5-(hydroxymethyl)oxolan-2-yl]-1,2,4-triazole-3-carboxamide | 451949 |
| 27. | Framycetin | 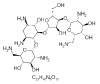 |
(2R,3S,4R,5R,6R)-5-amino-2-(aminomethyl)-6-[(1R,2R,3S,4R,6S)-4,6-diamino-2-[(2S,3R,4S,5R)-4-[(2R,3R,4R,5S,6S)-3-amino-6-(aminomethyl)-4,5-dihydroxyoxan-2-yl]oxy-3-hydroxy-5-(hydroxymethyl)oxolan-2-yl]oxy-3-hydroxycyclohexyl]oxyoxane-3,4-diol | 8378 |
In this present review, we provide insights on the sterility mosaic disease in pigeonpea (Cajanus Cajan) which causes devastating damage to the yield of pigeonpea. There was a major dilemma in etiology and molecular biology regarding the causal organism of SMD. The sterility mosaic virus is transmitted through the Aceria cajani eriophyid as a mite vector. Mite inoculates the whole field with the virus from their gut. Many preventive measures are being taken to overcome yield loss. Recently the identification was dependent on the symptoms, but characterization of the causal agent has manifest SMD detection by various techniques such as DAS-ELISA (A double antibody sandwich ELISA) and DIBA (A dot immunobinding assay) and many purification methods quasi-equilibrium zoning centrifugation Sucrose and CsCl gradients. PPSMV antibodies react with the virus in viruliferous mites seen in techniques such as DAS-ELISA and DIBA. Both of the techniques can detect the pathogen from the extracts of a minimum of 5 to 10 vector mites. The disease can be controlled by the various potential drugs such as Dicofol and 0.1% Fenazaquin as well as with molecular markers based on AFLP, SSR and SNPs markers. Beside genomics approach we can employ other approaches such as protein target interactions and chemoinformatics approach in this context few potential drugs against SMV which can provides stability and effectiveness towards the resistance in pigeonpea and which will also help in systematic agronomic system in diseased localities and exercise as a component in the integrated disease management.
NS acknowledges the funding from Department of Science and Technology, Government of India through DST INSPIRE Faculty Award Grant (DST/INSPIRE/04/2018/003674).
- Simtowe F, Shiferaw B, Kassie M, Abate T, Silim S, et al. Assessment of the current situation and future outlooks for the pigeonpea sub-sector in Malawi. Nairobi: ICRISAT. 2010.
- Bohra A, Saxena KB, Varshney RK, Saxena RK. Genomics-assisted breeding for pigeonpea improvement. Theoreti Appl Gene. 2020; 133: 1721-1737. PubMed: https://pubmed.ncbi.nlm.nih.gov/32062675/
- Singh N, Rai V, Singh NK. Multi-omics strategies and prospects to enhance seed quality and nutritional traits in pigeonpea. Nucleus. 2020; 63: 249–256.
- Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, et al. Draft genome sequence of pigeonpea (Cajanus cajan: an orphan legume crop of resource-poor farmers. Nat Biotechnol. 2012; 30: 83-89.
- Zavinon F, Adoukonou-Sagbadja H, Keilwagen J, Lehnert H, Ordon F, et al. Genetic diversity and population structure in Beninese pigeon pea [Cajanus cajan (L.) Huth] landraces collection revealed by SSR and genome wide SNP markers. Gene Resour Crop Evolut. 2020; 67: 191-208.
- Raisch MJ. Website and Database Survey 2016. J Int Econo Law. 2017; 20: 189-195.
- Mitra M. Report of the Imperial Mycologist. Report of the Imperial Mycologist. 1931.
- Kumar PL, Jones AT, Sreenivasulu P, Reddy DVR. Breakthrough in the identification of the causal virus of pigeonpea sterility mosaic disease. J Mycol Plant Pathol. 2000; 30.
- Jones AT, Kumar PL, Saxena KB, Kulkarni NK, Muniyappa V, et al. Sterility mosaic disease—the “green plague” of pigeonpea: advances in understanding the etiology, transmission and control of a major virus disease. Plant Dis. 2004; 88: 436-445. PubMed: https://pubmed.ncbi.nlm.nih.gov/30812645/
- Rani CS, Rani CSKS. Effect of different intercrops on growth and yield of pigeonpea in a paired row planting system. IJCS. 200; 8: 2258-2261.
- Kulkarni NK, Kumar PL, Muniyappa V, Jones AT, Reddy DVR. Transmission of Pigeon pea sterility mosaic virus by the eriophyid mite, Aceriacajani (Acari: Arthropoda). Plant Dis. 2002; 86: 1297-1302. PubMed: https://pubmed.ncbi.nlm.nih.gov/30818431/
- Patil BL, Dangwal M, Mishra R. Variability of Emaravirus species associated with sterility mosaic disease of pigeonpea in India provides evidence of segment reassortment. Viruses. 2017; 9: 183. PubMed: https://pubmed.ncbi.nlm.nih.gov/28696402/
- Saxena K, Bohra A, Choudhary AK, Sultana R, Sharma M, et al. The alternative breeding approaches for improving yield gains and stress response in pigeonpea (Cajanus cajan). Plant Breeding. 2021; 140: 74-86.
- Nawaz KA, David SM, Murugesh E, Thandeeswaran M, Kiran KG, et al. Identification and in silico characterization of a novel peptide inhibitor of angiotensin converting enzyme.pigeon pea (Cajanus cajan). Phytomedicine. 2017; 36: 1-7. PubMed: https://pubmed.ncbi.nlm.nih.gov/29157802/
- Dipshikha K, Seweta S, Chandra NB, Chauhan VB, Singh RN. Correlation between mite population (Aceriacajani) and environmental factors causing sterility mosaic disease of Pigeon pea. Int J Life Sci. 2013; 1: 228-232.
- Pedersen P, Grau C, Cullen E, Koval N, Hill JH. Potential for integrated management of soybean virus disease. Plant Dis. 2007; 91: 1255-1259. PubMed: https://pubmed.ncbi.nlm.nih.gov/30780527/
- Hill JH, Whitham SA. Control of virus diseases in soybeans. Adv Virus Res. 2014; 90: 355-390. PubMed: https://pubmed.ncbi.nlm.nih.gov/25410106/
- Elbeaino T, Digiaro M, Uppala M, Sudini H. Deep sequencing of pigeonpea sterility mosaic virus discloses five RNA segments related to Emaraviruses. Virus Res. 2014; 188: 27-31. PubMed: https://pubmed.ncbi.nlm.nih.gov/24685674/
- Sayiprathap BR, Patibanda AK, Kumari VP, Jayalalitha K, Rao V.S, et al. Prevalence of sterility mosaic disease (SMD) and variability in pigeonpea sterility mosaic virus (PPSMV) in southern-India. Indian Phytopathol. 2020; 1-10.
- Mielke-Ehret N, Mühlbach HP. Emaravirus: a novel genus of multipartite, negative strand RNA plant viruses. Viruses. 2012; 4: 1515-1536. PubMed: https://pubmed.ncbi.nlm.nih.gov/23170170/
- Kumar PL, Jones AT, Reddy DVR. A novel mite-transmitted virus with a divided RNA genome closely associated with pigeonpea sterility mosaic disease. Phytopathology. 2003; 93: 71-81. PubMed: https://pubmed.ncbi.nlm.nih.gov/18944159/
- Saxena KB, Lava Kumar P, Kulkarni NK, Jones AT, Muniyappa V, et al. Sterility mosaic disease the green plague of pigeonpea. Plant Dis. 2004; 88: 436-445.
- Singh NK, Gupta DK, Jayaswal PK, Mahato AK, Dutta S, Singh S, et al. The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol. 2012; 21: 98-112. PubMed: https://pubmed.ncbi.nlm.nih.gov/24431589/
- Kulkarni NK, Kumar PL, Muniyappa V, Jones AT, Reddy DVR. Transmission of Pigeon pea sterility mosaic virus by the eriophyid mite, Aceriacajani (Acari: Arthropoda). Plant Dis. 2002; 86: 1297-1302. PubMed: https://pubmed.ncbi.nlm.nih.gov/30818431/
- Saxena KB, Lava Kumar P, Kulkarni NK, Jones AT, Muniyappa V, et al. Sterility mosaic disease the green plague of pigeonpea. Plant Dis. 2004; 88: 436-445. PubMed: https://pubmed.ncbi.nlm.nih.gov/30812645/
- Latha TKS, Sabitha D. Interaction studies between pigeonpea sterility mosaic virus and mite vector. Indian J Plant Protection. 2004; 32: 105-110.
- Jones AT, Kumar PL, Saxena KB, Kulkarni NK, Muniyappa V, et al. Sterility mosaic disease—the “green plague” of pigeonpea: advances in understanding the etiology, transmission and control of a major virus disease. Plant Dis. 2004; 88: 436-445. PubMed: https://pubmed.ncbi.nlm.nih.gov/30812645/
- Gnanesh BN, Bohra A, Sharma M, Byregowda M, Pande S, et al. Genetic mapping and quantitative trait locus analysis of resistance to sterility mosaic disease in pigeonpea [Cajanus cajan (L.) Millsp.]. Field Crops Res. 2011; 123: 53-61.
- Latha TKS, Doraiswamy S. Detection of pigeonpea sterility mosaic virus, the causal agent of sterility mosaic disease of pigeonpea in viruliferous mite vector by DAS-ELISA and DIBA. Arch Phytopathol Plant Protect. 2008; 41: 537-541.
- Elbeaino T, Digiaro M, Uppala M, Sudini H. Deep sequencing of dsRNAs recovered.mosaic-diseased pigeonpea reveals the presence of a novel Emaravirus: pigeonpea sterility mosaic virus 2. Arch Virol. 2015; 160: 2019-2029. PubMed: https://pubmed.ncbi.nlm.nih.gov/26060057/
- Rajeswari E, Smitha KP, Kamalakannan A, Alice D, Bapu JK. Management of pigeonpea sterility mosaic disease. Legume Res. 2016; 39: 648-650.
- Pazhamala L, Saxena RK, Singh VK, Sameerkumar CV, Kumar V, Sinha P, et al. Genomics-assisted breeding for boosting crop improvement in pigeonpea (Cajanus cajan). Front Plant Sci. 2015; 6: 50. PubMed: https://pubmed.ncbi.nlm.nih.gov/25741349/
- Kumar PL, Latha TKS, Kulkarni NK, Raghavendra N, Saxena KB, et al. Broad‐based resistance to pigeonpea sterility mosaic disease in wild relatives of pigeonpea (Cajanus: Phaseoleae). Ann Appl Biol. 2005; 146: 371-379.
- Saxena RK, Kale S, Mir RR, Mallikarjuna N, Yadav P, et al. Genotyping-by-sequencing and multilocation evaluation of two interspecific backcross populations identify QTLs for yield-related traits in pigeonpea. Theoreti Appl Genet. 2020; 133: 737-749. PubMed: https://pubmed.ncbi.nlm.nih.gov/31844966/
- Sharma M, Rathore A, Mangala UN, Ghosh R, Sharma S, et al. New sources of resistance to Fusarium wilt and sterility mosaic disease in a mini-core collection of pigeonpea germplasm. Eur J Plant Pathol. 2012; 133: 707-714.
- Odeny DA, Githiri SM, Kimani PM. Inheritance of resistance to Fusarium wilt in pigeonpea {Cajanus cajan (L.) Millsp. 2009.
- Bhairappanavar SB, Fakrudin B, Rao KS. Inheritance Studies of Sterility Mosaic Disease (SMD) Resistance Cross Gullyalwhite×BSMR736 in Pigeonpea (Cajanus cajan (L.) Millsp.). Plant Gene Trait. 2014; 5.
- Raju NL, Gnanesh BN, Lekha P, Jayashree B, Pande S, et al. The first set of EST resource for gene discovery and marker development in pigeonpea (Cajanus Cajan L.). BMC Plant Biol. 2010; 10: 1-22.
- Singh S, Mahato AK, Jayaswal PK, Singh N, Dheer M, Goel P, et al. A 62K genic-SNP chip array for genetic studies and breeding applications in pigeonpea (Cajanus Cajan L. Millsp.). Sci Rep. 2020; 10: 1-14.
- Saxena KB, Lava Kumar P, Kulkarni NK, Jones AT, Muniyappa V, et al. Sterility mosaic disease the green plague of pigeonpea. Plant Dis. 2004; 88: 436-445. PubMed: https://pubmed.ncbi.nlm.nih.gov/30812645/
- Ujinwal M, Sahani PA, Singh N. Comparative Sequence and Structural Analysis of Lectin Protein in Chickpea (Cicer arietinum L.) and their Relationship with fabaceae Family. J Biomed Res Environ Sci. 2019; 5: 001-006.
- Mula MG, Saxena KB. Lifting the level of awareness on pigeonpea-a global perspective. International Crops Research Institute for the Semi-Arid Tropics. 2010.
- Kumar PL, Jones AT, Waliyar F. Biology, etiology and management of pigeonpea sterility mosaic. Ann Rev Plant Pathol. 2005; 3: 77-100.
- Kumar PL, Jones AT, Reddy DVR. Saxena KB, Lava Kumar P, et al. Sterility mosaic disease the green plague of pigeonpea. Plant Dis. 2003; 2004; 88: 436-445.
- Singh NK, Gupta DK, Jayaswal PK, Mahato AK, Dutta S, et al. The first draft of the pigeonpea genome sequence. J Plant Biochem Biotechnol. 2012; 21: 98-112. PubMed: https://pubmed.ncbi.nlm.nih.gov/24431589/
- Kulkarni NK, Kumar PL, Muniyappa V, Jones AT, Reddy DVR. Transmission of Pigeon pea sterility mosaic virus by the eriophyid mite, Aceriacajani (Acari: Arthropoda). Plant Dis. 2002; 86: 1297-1302. PubMed: https://pubmed.ncbi.nlm.nih.gov/30818431/
- Rajeswari E, Smitha KP, Kamalakannan A, Alice D, Bapu JK. Management of pigeonpea sterility mosaic disease. Legume Res. 2016 39: 648-650.
- https://pubchem.ncbi.nlm.nih.gov/compound/8268National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 6624, Chlorfenethol. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Chlorfenethol.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 26450, Proclonol. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Proclonol.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 14189996, 1-(4-Chlorophenyl)-1-phenylethanol. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/1-_4-Chlorophenyl_-1-phenylethanol.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 160701, 2,2-Dichloro-1,1-bis(4-chlorophenyl)ethanol. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/2_2-Dichloro-1_1-bis_4-chlorophenyl_ethanol.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 11348842. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/11348842.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 146570, Escitalopram. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Escitalopram.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 86287518. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Imidacloprid.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 47759, Furathiocarb. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Furathiocarb.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 54886, Benfuracarb. 2021.https://pubchem.ncbi.nlm.nih.gov/compound/Benfuracarb.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 47756. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/47756.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 3067994. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/3067994.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 131676698, Lactoferrin (322-329) (human). 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Lactoferrin-_322-329_-_human.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 456832. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/456832.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 6249, Ampicillin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Ampicillin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 5904, Penicillin g. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Penicillin-g.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 72493, Matromycin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Matromycin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 447043, Azithromycin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Azithromycin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 3779, Isoproterenol. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Isoproterenol.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 493570, Riboflavin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Riboflavin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 174174, Atropine. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Atropine.
- National Center for Biotechnology Information (2021). PubChem Substance Record for SID 349974776, Source: Therapeutic Target Database (TTD). 2021.https://pubchem.ncbi.nlm.nih.gov/substance/349974776.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 8378, Neomycin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Neomycin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 25074253. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/25074253.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 13903189. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/13903189.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 17856126, 1H-Diazepine-7-carboxylate. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/1H-Diazepine-7-carboxylate.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 44588235, Ningnanmycin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Ningnanmycin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 37542, Ribavirin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Ribavirin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 5090648, (+/-)-Tylophorine. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/5090648.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 92765, Cryptopleurine. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Cryptopleurine.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 639288. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/639288.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 100252, Ribavirin monophosphate. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Ribavirin-monophosphate.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 5064, Virazole. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Virazole.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 460516, Levovirin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Levovirin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 44443369, Tylophorinicine. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Tylophorinicine.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 36294, Tobramycin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Tobramycin.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 451447, Taribavirin hydrochloride. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Taribavirin-hydrochloride.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 451949. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/451949.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 8378, Neomycin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Neomycin\
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 16212126. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/16212126