More Information
Submitted: 04 June 2020 | Approved: 02 July 2020 | Published: 03 July 2020
How to cite this article: Kumari V, Kanwar SS. In silico comparative analysis of HIV protease inhibitors effect on 2019-nCoV coronavirus 3CLpro. Ann Proteom Bioinform. 2020; 4: 012-014.
DOI: 10.29328/journal.apb.1001011
Copyright License: © 2020 Kumari V, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: 2019-nCoV; Protease inhibitors; Pneumonia; 3CLpro; Interaction analysis; Ligands
In silico comparative analysis of HIV protease inhibitors effect on 2019-nCoV coronavirus 3CLpro
Vandna Kumari and Shamsher Singh Kanwar*
Department of Biotechnology, Himachal Pradesh University, Summer Hill, Shimla-171005. India
*Address for Correspondence: Shamsher S Kanwar, Department of Biotechnology, Himachal Pradesh University, Summer Hill, Shimla-171005, India, Tel/Fax: +91-177-2831948; Email: [email protected]
The novel coronavirus 2019-nCoV has become a bane to mankind and spread worldwide and infected many people. Thus, there is an urgent need of a cure for the severe pneumonia disease caused by this virus. In this study, In silico comparative analysis has been done for HIV protease inhibitors on coronavirus 3CLpro protein which has shown the major interactions and common amino acid residues involved in interactions. The amino acid interaction analysis has revealed two amino acids ARG4, LYS5 to be the major amino acids targets among selected ligands. The binding energy analysis has also revealed Cobicistat as one of these best suited ligand for 3CLpro.
The outbreak of novel coronavirus has caused major loss of human lives and many lives are still at risk. 2019-nCoV is an enveloped, positive-sense, single-stranded RNA beta-coronavirus [1]. Corona viruses can infect respiratory, gastrointestinal, hepatic and central nervous system of human [2], livestock, birds, bats [3], mouse, and many other wild animals [4]. Similar to SARS and MERS, non-structural proteins encoded by 2019-nCoV genome are 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRp) and helicase, structural proteins spike glycoprotein [1]. 3CLpro protein is considered as one of the main target for 2019-nCoV and the urgent need of cure has made it more prominent. In this study, the 3CLpro has been considered as a target and HIV protease inhibitors (ligands) were comparatively analyzed in order to reveal the interactions and important amino acids involved in such interactions, so as to make it easier to identify drug-targets.
Protein preparation
The three dimensional crystal structure of 2019-nCoV coronavirus protein 3CLpro (Pdb id: 6lu7) was obtained from RCSB PDB (https://www.rcsb.org/structure/). The ligand groups and water molecules already present were removed out in order to make this structure suitable for further interactions with other ligands.
Ligand preparation
The HIV protease inhibitors has been scanned for the selected coronavirus protein structure. Three ligands Lopinavir [5], Cobicistat [6], Amprenavir, Atazanavir, Indinavir and Saquinavir [7] were selected from the literature review. The chemical structures for these ligands were obtained from PubChem in SDF format and were converted to PDB format for further interactions analysis.
HIV protease inhibitors and 3CLpro interactions
The selected ligands were docked to coronavirus protein 3CLpro using Autodock tool 1.5.6 [8]. Autodock is an automated docking tool to predict how the small molecule or substrate bind to a specific target protein. The protein and ligand PDB formatted files were converted to PDBQT formats followed by docking using Lamarckian genetic algorithm and a total of ten conformations for each ligand docking were observed with their binding energies to find out the best lead compound among them. The protein ligand interactions were obtained followed by the identification of interacting amino acid residues of coronavirus protein 3CLpro to chemical groups with their respective ligands.
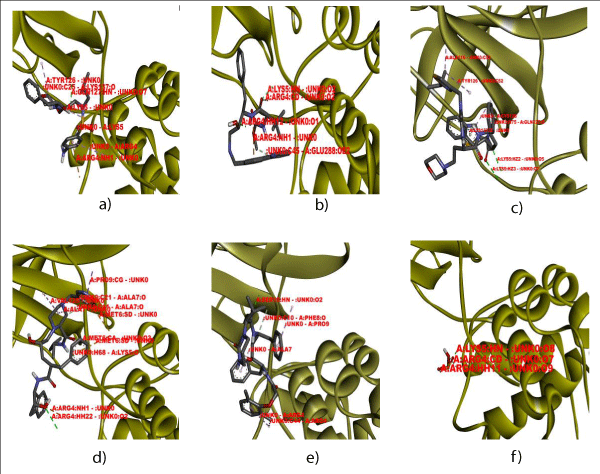
The docking analysis of 3CLpro and target ligands revealed the interaction types in three ligands. The docking studies involving 3CLpro with ligands such as Lopinavir, Cobicistat, Amprenavir, Atazanavir, Indinavir and Saquinavir were done to seek greater insight about the amino acid residues of MATE interacting with the specified ligands (Figure 1).
Figure 1: Interactions involved in docking of 3CLpro protein with ligands (a) Amprenavir (b) Atazanavir (c) Cobicistat (d) Indinavir (e) Lopinavir and (f) Saquinavur.
The interaction analysis revealed the occurrence of two amino acid residues ARG4 and LYS5 in interactions with selected ligands which conferred these two amino acid residues to be major drug target sites (Table 1).
| Table 1: Amino acid residues involved in interaction of 3CLpro to ligands. | |||
| Amino acids | Distance | Bond type | |
| Amprenavir | GLN127 | 2.69155 | Hydrogen bond |
| LYS137 | 3.38047 | Hydrogen bond | |
| ARG4 | 4.1211 | Electrostatic | |
| LYS5 | 5.46553 | Hydrophobic | |
| TYR126 | 4.82572 | Hydrophobic | |
| ARG4 | 5.4604 | Hydrophobic | |
| LYS5 | 5.33228 | Hydrophobic | |
| Atazanavir | ARG4 | 2.10606 | Hydrogen bond |
| LYS5 | 2.17532 | Hydrogen bond | |
| ARG4 | 3.7109 | Hydrogen bond | |
| GLU288 | 3.68988 | Hydrogen bond | |
| ARG4 | 3.88505 | Electrostatic | |
| Indinavir | ARG4 | 3.01365 | Hydrogen bond |
| LYS5 | 1.96033 | Hydrogen bond | |
| MET6 | 3.46698 | Hydrogen bond | |
| ALA7 | 3.41241 | Hydrogen bond | |
| ALA7 | 3.23456 | Hydrogen bond | |
| ARG4 | 3.41559 | Electrostatic | |
| PRO9 | 3.91164 | Hydrophobic | |
| MET6 | 5.23667 | Other | |
| MET6 | 5.90228 | Other | |
| ALA7 | 5.10413 | Hydrophobic | |
| VAL125 | 4.90336 | Hydrophobic | |
| Saquinavir | ARG4 | 1.91972 | Hydrogen bond |
| LYS5 | 1.91442 | Hydrogen bond | |
| ARG4 | 2.779 | Hydrogen bond | |
| Cobicistat | LYS5 | 2.82 | Hydrogen bond |
| LYS5 | 2.83 | Hydrogen bond | |
| GLN127 | 1.97 | Hydrogen bond | |
| LYS137 | 2.76 | Other | |
| ALA116 | 3.26 | Hydrophobic | |
| TYR126 | 4.84 | Hydrophobic | |
| CYS128 | 5.06 | Hydrophobic | |
| Lopinavir | SER10 | 2.04 | Hydrogen bond |
| PHE8 | 3.51 | Hydrogen bond | |
| ARG4 | 3.54 | Hydrophobic | |
| ALA7 | 4.64 | Hydrophobic | |
| ARG4 | 4.64 | Hydrophobic | |
| PRO9 | 4.64 | Hydrophobic | |
The binding energies for ligands and 3CLpro interactions were observed and analyzed comparatively. The lowest binding energy among the selected drugs; Cobicistat, Lopinavir, Amprenavir, Atazanavir, Indinavir and Saquinavir was found to be associated with Cobicistat which conferred it as one of the best suited ligand for 3CLpro (Table 2). However, Cobicistat lacks the enzyme inducing activity and thus requires the co-administered drugs with close monitoring of dose adjustments while treating the patients.
| Table 2: Binding energies for ligands docked with 2019-nCoV coronavirus 3CLpro protein. | |
| Ligand | Binding energy |
| Cobicistat | -2.41 |
| Amprenavir | -2.36 |
| Lopinavir | -2.33 |
| Indinavir | -2.01 |
| Atazanavir | -1.07 |
| Saquinavir | 1.16 |
The novel coronavirus 2019-nCoV has caused major loss to human lives, socially and economically as well. The urgent cure is the need of hour for this disease. The results observed from present studies has revealed the ligand with lowest binding energy ‘Cobicistat’ among the selected ligands which confers it to be one of the best suited ligand for 3CLpro. Cobicistat and Ritonavir are structural analogues of each other but Ritonavir has major limitations like poor water solubility which corresponds to difficulty in its co-formulation and drug interactions. Furthermore, the switching between Cobicistat-based drug and Ritonavir-based drug treatment may produce significant problems among the patients. In order to avoid these limitations, only cobicistat has been taken under consideration in this study. The common amino acids ARG4 & LYS5 in the docking studies confirmed these two amino acids as main targets. The amino acid residues involved in interaction with novel coronavirus protein 3CLpro also revealed the common interactions which shall further help scientists to find out the new target sites and lead compounds for novel coronavirus 2019-nCoV.
This work has been funded by the Department of Science & Technology, New Delhi, under a DST-INSPIRE Fellowship [IF180179] awarded to one of the authors (VK). The authors are thankful to DST and DBT, Ministry of Science & Technology, New Delhi for continuous financial support to the Bioinformatics Centre, Himachal Pradesh University, Shimla (India).
- Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov. 2020; 19: 149-150. PubMed: https://pubmed.ncbi.nlm.nih.gov/32127666/
- Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, et al. Isolation and characterization of a bat SARS‐like coronavirus that uses the ACE2 receptor. Nature. 2013; 503: 535‐538. PubMed: https://pubmed.ncbi.nlm.nih.gov/24172901/
- Wang LF, Shi Z, Zhang S, Field H, Daszak P, et al. Review of bats and SARS. Emerg Infect Dis. 2006; 12: 1834‐1840. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3291347/
- Chen Y, Guo D. Molecular mechanisms of coronavirus RNA capping and methylation. Virol Sin. 2016; 31: 3‐11. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7091378/
- Kim UJ, Won EJ, Kee SJ, Jung S, Jang HC. Combination therapy with lopinavir/ritonavir, ribavirin and interferon-alpha for Middle East respiratory syndrome. Antivir Ther. 2016; 21: 455-459. PubMed: https://pubmed.ncbi.nlm.nih.gov/26492219
- Pang J, Wang MX, Ang IYH, Tan SHX, Lewis RF, et al, Potential Rapid Diagnostics, Vaccine and Therapeutics for 2019 Novel Coronavirus (2019-nCoV): A Systematic Review J Clin Med. 2016; 9: 623. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7141113/
- Lv Z, Chu Y, Wang Y. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV AIDS. 2015;7: 95–104. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4396582/
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, et al. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J Comput Chem. 2009; 30: 2785-2791. PubMed: https://pubmed.ncbi.nlm.nih.gov/19399780/