Review Article
Microarray Analysis of Fish Genomic Data for enhancing Aquaculture Productivity of India

Ajit Kumar Roy*
Ex. Principal Scientist & Coordinator, Biotechnology Information Centre on Aquaculture of ICAR-CIFA Bhubaneswar), Prof. Institute of genetic Engineering, Badu, Kolkata, India
*Address for Correspondence: Ajit Kumar Roy, (Ex. Principal Scientist & Coordinator, Biotechnology Information Centre on Aquaculture of ICAR-CIFA Bhubaneswar), Prof. Institute of genetic Engineering, Badu, Kolkata, India, Tel: 09230011361; Email: [email protected]
Dates: Submitted: 05 August 2017; Approved: 25 August 2017; Published: 28 August 2017
How to cite this article: Roy AK. Microarray Analysis of Fish Genomic Data for enhancing Aquaculture Productivity of India. Ann Proteom Bioinform. 2017; 1: 006-017. DOI: 10.29328/journal.hpbr.1001002
Copyright License: © 2017 Roy AK. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Microarray; Fish genome; Aquaculture; Indian major carps; Salmon; Zebrafish
Abstract
This review gives a brief introduction to the microarray technology and its experimental design and data analysis and a discussion of recent global progress in research using microarray technology in fish biology and aquaculture. DNA microarrays have been reported to have been used for the analysis of gene expression during various physiological, developmental or cellular processes in fish. During the recent past, investigators have begun to use microarrays on fish to address ecological, evolutionary and environmental questions including the variability of gene expression in natural populations, speciation, ecotype diversity, environmental remediation and host-pathogen interactions. The study suggests that a lot of gene expression studies have been conducted on salmon and zebrafish in Europe and USA. The same may be applied on Indian Major Carps and Catfishes to augment productivity from aquaculture sector.
Introduction
A microarray is a collection of microscopic DNA spots attached to a solid surface. The DNA microarray is a tool used to determine whether the DNA from a particular individual contains a mutation in genes like BRCA1 and BRCA2.Now a days we use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Microarray technology has been applied to study of gene expression to study mechanisms of diseases and to accelerate the drug discovery process. There is a definite trend towards increasing the use of molecular diagnostic methods, and biochip technologies, along with bioinformatics techniques. Gene expression microarrays are tools that tell how much RNA (if any) a gene is making. Since 1977, and prior to microarray, only a few genes could be studied at a time using the northern blot analysis. Gene Chip microarrays use the natural chemical attraction, or hybridization, between DNA on the array and RNA target molecule from the sample based on complementary base pairs. Only RNA target molecule that have exact complementary base pair bind to the prob. Gene expression detection microarray is that they are able to measure tens of thousands of genes at a time, and it is this quantitative change in the scale of gene measurement that has led to a qualitative change in our ability to understand regulatory processes that occur at the cellular level. In other words, the short probe on the microarray measures the expression of the complete gene by sampling only a small section of the gene. In some instances, as little as one RNA molecule out of 100,000 different RNAs in an original sample may be detected. Microarray is one such technology is widely used by the researchers to investigate and address issues which were once thought to be nontraceable DNA microarrays are becoming a common tool in many areas of microbial research, including microbial physiology, pathogenesis, epidemiology, ecology, phylogeny, and pathway engineering. One can analyze the expression of many genes in a single reaction quickly and in an efficient manner. DNA Microarray technology helps in the identification of new genes and in knowing about their functioning and expression levels under different conditions, comparative genomics, and SNPs identification [1-6]. Proteome chips have been used to study protein-protein interactions [7], protein-DNA interactions [8], protein-lipid interactions [7], protein-drug interactions [9], protein-receptor interactions [10], and antigen-antibody interactions [11]. In addition, proteome chips have been used to study kinase activities [7,12] and have been used for serum profiling [13]. This chapter has focused on use of microarray technology in understanding biological process, and its subsequent applications in Fish biology aiming to enhance aquaculture productivity.
Genomic technology and post-genome technology
The advent of micro-array technology revolutionized biomedical research, for the first time allowing overall characterization (genome-wide) of gene expression. Yet the technology had several flaws limiting sensitivity and specificity that are overcome by sequencing. A first application of NGS was in gene expression profiling. Hybridization on DNA microarrays is a proven technique in genomics research because of its. Flexibility and low cost of production. Microarrays with low-density probes have been used in single-nucleotide polymorphism detection as well as nucleic acid diagnostic application. The impact of microarrays on the development of these new tools have started modifying our daily practice of experimental
How does a DNA microarray work?
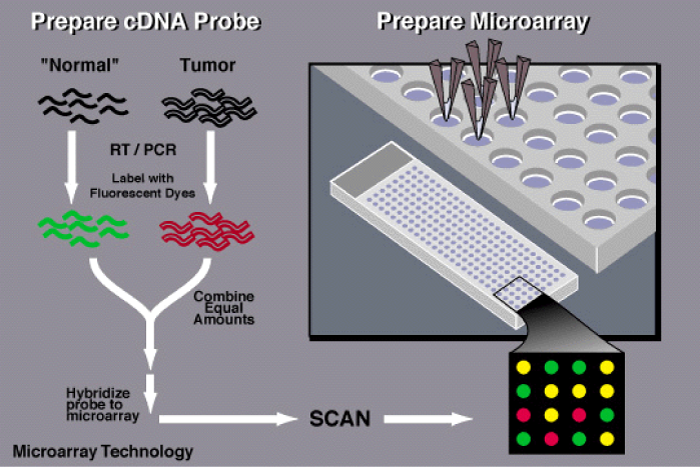
Figure 1 shows research in biology. Application of new technology like DNA and protein microarrays necessitates input from molecular biologists, geneticists, chemists, physicists, mathematicians, computer scientists, and engineers. Protein based microarrays are key to this goal, permitting both the characterization of disease systems and the system-wide pharmacophoric properties of putative therapeutic agents. Many cutting-edge microarray analysis tools and algorithms, including commonly used limma and affy packages in Bioconductor, need sophisticated knowledge of mathematics, statistics and computer skills for implementation. Commercially available software can provide a user-friendly interface at considerable cost. To facilitate the use of these tools for microarray data analysis on an open platform is available that is developed an online microarray data analysis platform, WebArray, for bench biologists to utilize these tools to explore data from single/dual color microarray experiments. The currently implemented functions were based on limma and affy package from Bioconductor, the spacings LOESS histogram (SPLOSH) method, PCA-assisted normalization method and genome mapping method. Web Array incorporates these packages and provides a user-friendly interface for accessing a wide range of key functions of limma and others, such as spot quality weight, background correction, graphical plotting, normalization, linear modeling, empirical Bayes statistical analysis, false discovery rate (FDR) estimation, chromosomal mapping for genome comparison. Web Array offers a convenient platform for bench biologists to access several cutting-edge microarray data analysis tools. The website is freely available at http://bioinformatics.skcc.org/webarray [14].
Figure 1: To determine whether an individual possesses a mutation for a particular disease, a scientist first obtains a sample of DNA from the patient's blood as well as a control sample-one that does not contain a mutation in the gene of interest.
Application of the microarray technology in the studies of Fish Biology
If aquaculture is to play a vital role in ensuring future fish availability for food security and nutrition, this sector has to adopt best practice in aquaculture that requires a full understanding of the genomic controls and transcriptional profiles of cultured fish species as well as improvements in aquaculture through regulation of the expression of functional genes. In the post genomic era, the use of DNA microarray technology in fish biology and aquaculture may have great significance and may be applied to discovery of novel genes, gene expression profiling from fish species of interest, and identification of the genomic responses to environmental stimulation in aquaculture. The use of microarrays for the study of various aspects of fish physiology has seen a spectacular increase in recent years from model species, such as zebra fish, to current studies with commercially important species, such as salmonids, catfish, carp, and flatfish. Besides micro array technology has emerged as a key tool for understanding developmental processes as well as basic physiology. Recent development is the application of micro arrays technology in the fields of ecotoxicology and nutrigenomics. Microarray and gene expression analysis have been key in our understanding of molecular pathways underlying physiological responses. DNA microarrays have been used for the analysis of gene expression during various physiological, developmental or cellular processes in fish. During the recent past, investigators have begun to use microarrays on fish to address ecological, evolutionary and environmental questions including the variability of gene expression in natural populations, speciation, ecotype diversity, environmental remediation and host-pathogen interactions. Given their commercial importance and the availability of several microarray platforms, most of the research in these areas has focused on salmonids. However, the same issues could be studied in a number of fish species like Indian Major Carps, catfish, prawn and minor carps of commercial interest and nutritional values.
As established by quantitative RT-qPCR before the advent of microarrays, 4 to 8 days after DNA vaccination by intramuscular injection, gene expression by fish hematopoietic organs showed an increase in interferon-inducible mx [15-20], virally induced genes (Vig) [21,22] and mhc and tcr genes [23]. The recent availability of fish microarrays [24], which allow the expression profiling of thousands of genes simultaneously, has provided new opportunities to further study fish immunological responses in several rhabdovirus/fish models. Traditional sequencing, annotation and estimation of frequencies of each rhabdovirallyinduced transcript in flatfish, is one of the strategies designed to identify genes transcribed after rhabdoviral infections (pathogen-induced gene approach) [25]. To identify HRV-induced genes were made in the Japanese flounder Paralichthys olivaceous by sequencing 300-596 expressed sequence tag (EST) clones from leukocytes 2-5 days after infection. The frequencies of each EST were estimated within a short 1 to 10 range [26,27]. Japanese flounder EST-derived cDNA microarrays were applied to in vitro kidney cell cultures 3-6 h after HRV infection [28].
Recently, many studies have used microarray technology to determine gene profiles in different fish species. A microarray studies on various species of fishes have been reported [29-37] Literature review suggests that in India few studied have been conducted on Indian major carps [38-40]. The use of DNA microarray technology in fish biology and aquaculture may have great significance and may be applied to discovery of novel genes, gene expression profiling from fish Species of interest, and identification of the genomic responses to environmental stimulation in aquaculture.
Use of the microarray technology in the studies of model Fish, specifically on Zebra Fish and Salmon Fish
Microarrays have already been used to quantify fish gene expression as well as to discover new genes involved in defense in several fish rhabdovirus models, such as flatfish, salmonid (salmon and trout) and zebrafish. Genes that show increased transcription after infection (hypothetically signaling internal organs to react against the viral invasion) and genes whose transcription is inhibited (possibly due to viral shut-off of critical host defenses) might help researchers in their quest to identify new adjuvant candidates for fish vaccines. Few studies have used microarray technology to determine gene profiles in different fish species [5]. A microarray consisting of 4512 complementary DNAs (cDNAs) was constructed to investigate the adaptive molecular responses of zebra fish to hypoxia during development [6,7]. Linney et al. [8], analyzed 15512 unique transcripts from wild Danio rerio and found that 23 muscle specific genes expression data is elucidated, however, it is much analyzed 15512 unique transcripts from wild-type challenging.
Expressed sequence tag (EST)-based microarrays of the Japanese flounder, trout, salmon and zebrafish have been used in gene-discovery efforts. These studies included infections with IHNV [41,42], VHSV [43-45] and hirame rhabdovirus (HRV) [46].The expression of a number of unknown genes was also increased [25]. Among these, the LB3 [8] gene increased a maximum of 56-fold 3 days after infection and then remained increased for one week [43]. Microarrays in the study of the salmonid/IHNV/VHSV models Large-scale genomic projects for salmon have been initiated by groups in Canada, the USA, the UK, Norway and France. As a result there are many physical and genetic maps, large collections of ESTs and a growing number of genomic sequences and derived microarrays. Thus three projects have developed salmonid microarrays. The first salmonid 16K cDNA microarray appeared in 2004. This array was developed by the Genomic Research on Atlantic Salmon Project (GRASP) [47] and led to the most recent 32K cDNA [48,49] and the first 5K oligo DNA of 70-mer [50] microarrays. The high sequence similarity (~86 %) between salmonids (9 genera and 68 species) indicates that cDNA microarrays may be suitable for studies involving any member of this fish family. Transcriptome Analysis of Important Traits of Salmon (TRAITS) and the Norwegian Salmon Genome Project (SGP) also developed a 16K cDNA microarray (http://www.abdn.sfirc/salmon) based on two independent collections of their bacterial clones kept in ARK, Genomics Facility at Roslin Institute, UK and at SGP Genetics Laboratory at the University of Oslo, respectively. The TRAITS-SGP cDNA array was obtained from ESTs from 15 tissues (pathogen-induced libraries, trait-specific subtractive EST, starvation-induced libraries, diet-response libraries, smoltification-response libraries and well-known genes). This array was conceived as a preliminary tool to develop an oligo microarray for routine health monitoring of Atlantic salmon. The first results found some artefactual expression patterns caused by cross-hybridization of similar transcripts and underlined the greater relevance of biological over technical replicates [51].
The new rainbow trout (Oncorhynchus mykiss) high-density, oligonucleotide microarray was developed using 37394 specific 60-mer oligonucleotide probes assembled from 244984 ESTs from 12 tissues (http://compbio.dfci.harvard.edu/tgi/cgibin/tgi/gimain.pl?gudb¼r_trout). The specificity of each probe was checked for possible non-specific mRNA cross-hybridization by comparing all individual probes with all rainbow trout transcriptome sequences. Approximately 91 % of the sequences used for this microarray matched a previously annotated sequence in the GenBank. Few attempts have been made to use these microarrays to study the rhabdoviral immunization of salmonids. In homozygous trout, the 16 K cDNA GRASP microarray was used to profile 7-day muscle transcripts after intramuscular injection of the IHNV G gene [47,49]. After immunization, irf3, mx, vig1, and vig8 transcripts were increased [42]. Genes associated with antigen presenting cells, lymphocytes, leukocytes, inflammation, antigen presentation, and interferon pathways were also augmented. The increased levels of transcripts associated with type I IFN pathways in systemic organs (gill, spleen and kidney) were corroborated by RT-qPCR. These observations confirmed that, when intramuscularly injected, the host expressed viral G gene induces a systemic non-specific type 1 IFN innate immune response. Using a 1.8K cDNA salmonid microarray, comparison of infection-by-injection with IHNV and attenuated IHNV in rainbow trout after 1 and 3 days showed an IHNV-dependent change in differential transcription in kidney towards adaptive immunity genes. Thus, the rapid spread of the IHNV infection inhibited tnfa, mhc1, and several other gene markers while favouring mhc2 and ig responses. The molecular mechanism for the development of late (months) specific cytotoxic T or B cell-mediated humoral responses has not been addressed by means of microarrays [52,53]. More recently, trout families with low (32% survival following challenge) and high susceptibility to VHSV (18% survival following challenge) were infected with VHSV by bath exposure and transcriptional data from internal organs were analyzed with the 16K GRASP microarray from day 3 post-challenge [54]. In total, 939 genes were differentially expressed between infected and non-infected fish. The genes increased in infected fish belonged to the following categories: stress and defense response, NFkappaB signal transduction, response to non-self, antigen processing and presentation, and proteasome complexes. Most were also increased among the 642 differentially expressed genes in the low-susceptibility trout family but not among the 556 differentially expressed genes in the high-susceptibility family. These results suggest that the innate immune system of internal organs plays a crucial role in eliciting an effective immune response to VHSV infection in rainbow trout [54].
The zebrafish Danio rerio is one of the most suitable models in which to carry out microarray studies because, compared to other fish, its genome sequence is one of the most advanced. Furthermore, ~ 40 K annotated quantitative polymerase chain reaction (qPCR) arrays and annotated oligo microarrays are available. In addition, large-scale experimentation with zebrafish is easier than with other fish models and zebrafish are susceptible to several viruses, most of these belonging to the fish rhabdoviral family [55]. Of these, VHSV [56] was chosen in a recent study using microarrays [57] over IHNV [58], snake-head rhabdovirus (SHRV) [59] and spring viremia of carp (SVC) [60], because only in the VHSV/zebrafish model have infection-by-immersion (the natural route of infection) and successful vaccination been described [56]. Damage and epithelial cell death immediately after VHSV infection in the surface portals of entry of these viruses, such as the fins [61], should alert surrounding cells to promote epithelial cell division to replace dead cells, recruit inflammatory cells to the infection site, and send signals to internal immune organs. However, viral-induced signals to inhibit the most relevant host responses have also been detected. Detection of natural early responses may contribute to identifying vaccine adjuvants. Thus, the expression of the 636 immune-related transcripts that were increased after VHSV infection, as estimated by hybridization to oligo microarrays (confirmed by RT-qPCR arrays), was higher in fins than in organs. In contrast, the number of decreased transcripts was higher in organs than in fins. Therefore, an upregulated response of immune-related genes was greatest in fin tissues, while a downregulated response was most detected in the internal organ responses. The latter might be targets of viral inhibitory signals early after infection [57]. These results showed that 2 days after infection-by-immersion, VHSV had not yet caused a strong response from zebrafish internal organs, which contrasts with reports in other fish at later times after infection-by-injection (such as ifn1, mx, il1b, tnfa, etc.) [62-65] or infection-by-immersion [54,66]. The zebrafish are refractory to rhabdoviral infection-by-immersion at high temperatures or without acclimatization to low temperatures with IHNV [58], VHSV [56] or SVC [60]. So, a temperature-dependent response mechanism(s) that inhibits rhabdoviral infection and spread may occur. While these preliminary findings shed some light on the earliest effects of VHSV infection at the molecular level, some of the new immune-related genes identified might be suitable candidate adjuvants for fish vaccines [67-69]. To best detect innate immune responses, early times after rhabdoviral infection should be studied. Thus, according to the data obtained from flatfish, salmonid and zebrafish studies, the maximal number of >2-fold differentially expressed genes in microarrays was detected 2-3 days after rhabdoviral infection [26,43,44, 6,18,27]. As with many other viruses and host species, an increase in ifn1 expression is one of the first responses to the injection of any DNA vaccine [15] and to rhabdoviral infections [70]. Transcripts encoding several forms of il17 were detected as differentially expressed only in one of the studies using microarrays [57].
Development of a 37-K high-density oligonucleotide microarray
A new tool for functional genome research in rainbow trout: A rainbow trout high-density oligonucleotide microarray was constructed using all tentative consensus (TC) sequences that are publicly available from all international rainbow trout Oncorhynchus my kiss genomic research projects through the Rainbow Trout Gene Index database. The new array contains 60-mer oligonucleotide probes representing 37 394 unique TC sequences and 1417 control spots. The array (4×44 format) was manufactured according to the design by Agilent Technologies using the inkjet-based Sure Print technology (design number 016320). The performance of the new microarray platform was evaluated by analyzing gene expression associated with rainbow trout, Vitello genesis-induced muscle atrophy. This microarray will open new avenues of research that will aid in the development of novel strategies for genetic improvement for economically important traits benefiting the salmonid aquaculture industries [72].
Expansion of the Genomics Research on Atlantic salmon Salmo Salar L. Project (GRASP) microarray tools
Salmonids are the most widely studied group of fish, and in the last few years, genomics technologies have begun to contribute to this rich biology. The first salmonid microarrays appeared in 2004 and since then several dozen studies have demonstrated the utility of genomic approaches. The widespread use of the genomics research on Atlantic salmon project 16 k array and greatly expanded genome resources have led to the development of an experimental 5 k oligo (70-mer) array and a 32 k cDNA microarray in the near future. In this paper, the authors examined some of the procedures used in the development of past arrays and reexamined them in light of new genomic data available. Some preliminary control experiments of the new 5 k array were investigated that examine oligo designs based on distance from the polyA tail, the effects of mismatches and cross-species hybridization specificity. Beneficial approaches are also identified in the development of the new 32 k cDNA array [47], and cDNA microarray studies on Atlantic salmon [51] and on Cyprinus carpio [73]. Hepatic gene expression profiles in juvenile rainbow trout (Oncorhynchus my kiss) fed fishmeal or fish oil-free diets is reported [74]. It is observed that seventy-one and seventy-five genes were affected after fish oil and fishmeal replacement respectively. The major part of modified gene expression coding for proteins of the metabolic pathways was as follows: (i) a lower level of expression for genes of energy metabolism found in fish after fishmeal and fish oil replacement; (ii) a lower level of gene expression for fatty acid metabolism (biosynthesis) in fish fed with vegetable oils; (iii) a differential expression of actors of detoxification metabolism in trout fed with vegetable oils; (iv) a lower level of expression of genes involved in protein metabolism in fish fed with plant proteins. Overall, our data suggest that dietary fish oil replacement is linked to a decreased capacity of fatty acid biosynthesis (fatty acid syntheses) and variation of detoxification metabolism (cytochrome P450s) whereas dietary fishmeal replacement may depress protein metabolism in the liver as reflected by glutamine synthesize.
Use of microarray technology in the studies of fish reproduction
Several articles are available on Steroid-induced sex reversal in rainbow trout is reported [75]. Intra-gonadal molecular mechanisms involved in the regulation of rainbow trout spermatogenesis is studied [76]; Hormonal induction or photoperiod manipulation of ovulation result in differences in the egg maternal mRNA abundance of specific genes-Prohibitin mRNA accumulation is negatively correlated to embryo developmental potential-The hormonal induction and the photoperiod manipulation of the ovulation affect egg quality through common and distinct mechanisms [78,79]; Molecular phenotype of zebrafish ovarian follicle by serial analysis of gene expression and proteomic profiling, and comparison with the transcriptomes of other animals [80]; A comprehensive survey of the genes involved in maturation and development of the rainbow trout ovary [81] and Sensitivity issues in DNA array-based expression measurements and performance of nylon microarrays for small samples [82]. Identification of candidate’s genes involved in oocyte maturation and/or ovulation [83]; Identification of proteins in full grown oocytes [77] Identify new proteins that may have an extra-ovarian origin-Identify new proteins may have been accumulated much before gene expression ends and identification of proteins in body fluids -search for protein markers of post-ovulatory egg ageing in trout coelomic fluid [84]. Use of a generic cDNA microarray to study rainbow trout reproductive issues and Sex ratio control-natural sex differentiation and steroid induced sex reversal [75]; Control of male sexual maturation-spermatogenesis regulation by gonadotropins and sex steroid hormones [76]. Hormonal or photoperiod control of ovulation affect egg quality ([77] maternal inherited RNA stocks in full grown egg of zebrafish [79]; Oogenesis and egg quality-Follicular competence to ovulation -Egg quality after natural or controlled ovulation [79]).
Conclusion
The study indicates that a lot of gene expression studies have been conducted on salmon and zebrafish in Europe and USA. The same may be applied on Indian Major Carps and Catfishes to augment productivity from aquaculture sector it is expected that abovementioned compilation/findings will be helpful for future researchers in perusing studies on species of Indian origin.
Acknowledgements
The author is thankful to the unanimous referee for suggestions in improving the manuscript.
References
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996; 14: 457-460. Ref.: https://goo.gl/DQP4U2
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995; 270: 467-470. Ref.: https://goo.gl/JiU8bG
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999; 21: 10-14. Ref.: https://goo.gl/oDXwCx
- Heller RA, Schena M, Chai A, Shalon D, Bedilion T, et al. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci USA. 1997; 94: 2150-2155. Ref.: https://goo.gl/wZxu5c
- Affymetrix GeneChip technology. An overview of how an Affimatrix GeneChip is made. Ref.: https://goo.gl/PBo4Ab
- Crowther DJ. Applications of microarrays in pharmaceutical industry. Curr Opin Pharmacol. 2002; 2: 551-554. Ref.: https://goo.gl/9X5U82
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, et al. Global analysis of protein activities using proteome chips. Science. 2001; 293: 2101-2105. Ref.: https://goo.gl/Wo7btP
- Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, et al. Regulation of gene expression by a metabolic enzyme. Science. 2004; 306: 482-484. Ref.: https://goo.gl/cVkP9v
- Huang J, Zhu H, Haggarty SJ, Spring DR, Hwang H, et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc Natl Acad Sci U S A. 2004; 101: 16594-16599. Ref.: https://goo.gl/P1Vvfn
- Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006; 439: 168-174. Ref.: https://goo.gl/hh81jm
- Michaud GA, Salcius M, Zhou F, Bangham R, Bonin J, et al. Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol. 2003; 21: 1509-1512. Ref.: https://goo.gl/tjvNDv
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005; 438: 679-684. Ref.: https://goo.gl/NT2oPP
- Zhu H, Hu S, Jona G, Zhu X, Kreiswirth N, et al. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc Natl Acad Sci U S A. 2006; 103: 4011-4016. Ref.: https://goo.gl/kMAKKz
- Aittokallio T, Kurki M, Nevalainen O, Nikula T, West A, et al. Computational strategies for analyzing data in gene expression microarray experiments. J Bioinform Comput Biol. 2003; 1: 541-586. Ref.: https://goo.gl/GsV1sX
- Acosta F, Collet B, Lorenzen N, Ellis A. E. Expression of the glycoprotein of viral haemorrhagic septicemia virus (VHSV) on the surface of the fish cell line RTG-P1 induces type 1 interferon expression in neighboring cells. Fish Shellfish Immunol. 2006; 21: 272-278. Ref.: https://goo.gl/Cffg8Q
- Boudinot P, Blanco M, de Kinkelin P, Benmansour A. Combined DNA immunization with the glycoprotein gene of viral hemorrhagic septicemia virus and infectious hematopoietic necrosis virus induces double-specific protective immunity and nonspecific response in rainbow trout. Virology. 1998; 249: 297-306. Ref.: https://goo.gl/8tngfC
- McLauchlan PE, Collet B, Ingerslev E, Secombes CJ, Lorenzen N, et al. DNA vaccination against viral haemorrhagic septicemia (VHS) in rainbow trout: size, dose, route of injection and duration of protection-early protection correlates with Mx expression. Fish Shellfish Immunol. 2003; 15: 39-50. Ref.: https://goo.gl/QBYYdh
- Purcell MK, Kurath G, Garver KA, Herwig RP, Winton JR. Quantitative expression profiling of immune response genes in rainbow trout following infectious haematopoietic necrosis virus (IHNV) infection or DNA vaccination. Fish Shellfish Immunol. 2004; 17: 447-462. Ref.: https://goo.gl/CQRtUQ
- Robertsen B. Expression of interferon and interferon-induced genes in salmonids in response to virus infection, interferon-inducing compounds and vaccination. Fish Shellfish Immunol. 2008; 250: 351-357. Ref.: https://goo.gl/kGNWxg
- 20.Taggart JB, Bron JE, Martin SAM, Seear PJ, Hoyheim B, et al. A description of the origins, design and performance of the TRAITS-SGP Atlantic salmon Salmo salar L. cDNA microarray. J Fish Biol. 2008; 72: 2071-2094. Ref.: https://goo.gl/xVDJnC
- Boudinot P, Massin P, Blanco M, Riffault S, Benmansour A. vig-1 a new fish gene induced by the rhabdovirus glycoprotein, has a virus-induced homologue in humans and shares conserved motifs with the MoaA family. J Virol. 1999; 73: 1846-1852. Ref.: https://goo.gl/N5g1bF
- Boudinot P, Salhi S, Blanco M, Benmansour A. Viral haemorrhagic septicaemia virus induces vig-2, a new interferon-responsive gene in rainbow trout. Fish Shellfish Immunol. 2001; 11: 383-397. Ref.: https://goo.gl/TihrWb
- Takano T, Iwahori A, Hirono I, Aoki T. Development of a DNA vaccine against hirame rhabdovirus and analysis of the expression of immune-related genes after vaccination. Fish Shellfish Immunol. 2004; 17: 367-374. Ref.: https://goo.gl/anuZ72
- Martin SAM, Collet B, Mackenzie S, Evensen O, Secombes CJ. Genomic tools for examining immune gene function in salmonid fish. Reviews on Fisheries Science. 2008; 16: 112-118. Ref.: https://goo.gl/K6KQNP
- Aoki T, Hirono I, Kondo H, Hikima J, Jung TS. Microarray technology is an effective tool for identifying genes related to the aquacultural improvement of Japanese flounder, Paralichthys olivaceus. Comarative Biochemical Physiology Part D Genomics Proteomics. 2011; 6: 39-43. Ref.: https://goo.gl/6TCvaR
- Aoki T, Nam BH, Hirono II, Yamamoto E. Sequences of 596 cDNA Clones (565,977 bp) of Japanese Flounder (Paralichthys olivaceous) Leukocytes Infected with Hirame Rhabdovirus. Marine Biotechnology. 1999; 1: 477-488. Ref.: https://goo.gl/BDBFWL
- Nam BH, Yamamoto E, Hirono I, Aoki T. A survey of expressed genes in the leukocytes of Japanese flounder, Paralichthys olivaceus, infected with Hirame rhabdovirus. Dev Comp Immunol. 2000; 24: 13-24. Ref.: https://goo.gl/JSQM5w
- Kurobe T, Yasuike M, Kimura T, Hirono I, Aoki T. Expression profiling of immune-related genes from Japanese flounder Paralichthys olivaceus kidney cells using cDNA microarrays. Dev Comp Immunol. 2005; 29: 515-523. Ref.: https://goo.gl/MjEPi7
- Woo Y, Affourtit J, Daigle S, Viale A, Johnson K, et al. A comparison of cDNA, oligonucleotide, and Affymetrix Gene Chip gene expression microarray platforms. J BiomolTech. 2004; 15: 276-284. Ref.: https://goo.gl/M4Anc7
- Hoffmann JL, Torontali SP, Thomason RG, Lee DM, Brill JL, et al. Hepatic gene expression profiling using gene chips in zebra fish exposed to 17alpha-ethynylestradiol. Aquat Toxicol. 2006; 79: 233-246. Ref.: https://goo.gl/Z5Jv4F
- Moens LN, van der Ven K, Van Remortel P, Del-Favero J, De Coen WM. Expression profiling of endocrine disrupting compounds using a customized cyprinus carpio cDNA microarray. Toxicol Sci. 2006; 93: 298-310. Ref.: https://goo.gl/LbKWeM
- Hook SE, Skillman AD, Small JA, Schultz IR. Gene expression patterns in rainbow trout, Oncorhynchus my kiss, exposed to a suite of model toxicants. Aquat Toxicol. 2006; 77: 372-385. Ref.: https://goo.gl/w4xdKZ
- Gunnarsson L, Kristiansson E, Forlin L, Nerman O, Joakim Larsson DG. Sensitive and robust gene expression changes in fish exposed to estrogen-a microarray approach. BMC Genomics. 2007; 8: 149. Ref.: https://goo.gl/fJpvZT
- William Goetz F, MacKenzie S. Functional genomics with microarrays in fish biology and fisheries. Fish and Fisheries. 2008; 9: 378-395. Ref.: https://goo.gl/1QByWu
- Ayaka Y, von Schalburg K, Cooper G, Koop BF, Yoshizaki G. Identification of a molecular marker for type A spermatogonia by microarray analysis using gonadal cells from p vasa-GFP transgenic rainbow trout (Oncorhynchus mykiss). Molecular Reproduction and Development. 2009; 76: 246-254. Ref.: https://goo.gl/yCmx2L
- Castilho PC, Buckley BA, Somero G, Barbara AB. Heterologous hybridization to a complementary DNA microarray reveals the effect of thermal acclimation in the endothermic blue fin tuna (Thunnus orientalis). Molecular Ecology. 2009; 18: 2092-2102. Ref.: https://goo.gl/X7ELSU
- Zhang J, Chu W, Guihong Fu. DNA microarray technology and its application in fish biology and aquaculture. Frontiers of Biology in China. 2009; 4: 305-313. Ref.: https://goo.gl/je8YUU
- Roy AK, Martha SR. Correspondence Analysis in Study of Genomic Sequence of Labeo rohita. In: Applied Bioinformatics, Statistics and Economics in Fisheries Research. 2008; 29-47. Ref.: https://goo.gl/EoDuFZ
- Roy AK, Sahu CK, Martha SR. Statistical Design and Analysis of Microarray Data of Carps. In: Applied Bioinformatics, Statistics and Economics in Fisheries Research. 2008; 49-72. Ref.: https://goo.gl/aDAQJQ
- Roy AK, Martha SR. Statistical Methods for Genomic Sequence and Microarray Analysis. Proc. National Workshop on ‘Molecular Modeling and protein docking held during. 2008; 3-17.
- MacKenzie S, Balasch JC, Novoa B, Ribas L, Roher N, et al. Comparative analysis of the acute response of the trout, O. mykiss, head kidney to in vivo challenge with virulent and attenuated infectious hematopoietic necrosis virus and LPS-induced inflammation. BMC Genomics. 2008; 26: 141. Ref.: https://goo.gl/Jw6mUU
- Purcell MK, Nichols KM, Winton JR, Kurath G, Thorgaard GH, et al. Comprehensive gene expression profiling following DNA vaccination of rainbow trout against infectious hematopoietic necrosis virus. Mol Immunol. 2006; 43: 2089-2106. Ref.: https://goo.gl/Bk3QFf
- Byon JY, Ohira T, Hirono I, Aoki T. Use of a cDNA microarray to study immunity against viral hemorrhagic septicemia (VHS) in Japanese flounder (Paralichthys olivaceus) following DNA vaccination. Fish Shellfish Immunol. 2005; 18: 135-147. Ref.: https://goo.gl/jkRggW
- Byon JY, Ohira T, Hirono I, Aoki T. Comparative immune responses in Japanese flounder, Paralichthys olivaceous after vaccination with viral hemorrhagic septicemia virus (VHSV) recombinant glycoprotein and DNA vaccine using a microarray analysis. Vaccine. 2006; 24: 921-930. Ref.: https://goo.gl/TR7UYy
- Encinas P, Rodriguez-Milla MA, Novoa B, Estepa A, Figueras A, et al. Zebrafish fin immune responses during high mortality infections with viral haemorrhagic septicemia rhabdovirus. A proteomic and transcriptomic approach. BMC Genomics. 2010; 11: 518-534. Ref.: https://goo.gl/GSCW7t
- Fernandez-Alonso M, Rocha A, Coll JM. DNA vaccination by immersion and ultrasound to trout viral haemorrhagic septicemia virus. 2001; 19: 3067-3075. Ref.: https://goo.gl/7hekdp
- Yasuike M, Kondo H, Hirono I, Aoki T. Difference in Japanese flounder, Paralichthys olivaceous gene expression profile following hirame rhabdovirus (HIRRV) G and N protein DNA vaccination. Fish Shellfish Immunol. 2007; 23: 531-541. Ref.: https://goo.gl/dfJub4
- von Schalburg KR, Rise ML, Brown GD, Davidson WS, Koop BF. A comprehensive survey of the genes involved in maturation and development of the rainbow trout ovary. Biological Reproduction. 2005; 72: 687-699. Ref.: https://goo.gl/JiDVYN
- von Schalburg KR, Cooper GA, Leong J, Robb A, Lieph R, et al. Expansion of the genomics research on Atlantic salmon Salmo salar L. project (GRASP) microarray tools. J Fish Biol. 2008; 72: 2051-2070. Ref.: https://goo.gl/H5gf6z
- von Schalburg KR, Rise ML, Cooper GA, Brown GD, Gibbs AR, et al. Fish and chips: various methodologies demonstrate utility of a 16,006-gene salmonid microarray. BMC Genomics. 2005; 6: 126-136. Ref.: https://goo.gl/d53491
- Koop BF, von Schalburg KR, Leong J, Walker N, Lieph R, et al. A salmonid EST genomic study: genes, duplications, phylogeny and microarrays. BMC Genomics. 2008; 9: 545-555. Ref.: https://goo.gl/iDFjuy
- Taggart JB, Bron JE, Martin SAM, Seear PJ, Hoyheim B, et al. A description of the origins, design and performance of the TRAITS-SGP Atlantic salmon Salmo salar L. cDNA microarray. J Fish Biol. 2008; 72: 2071-2094. Ref.: https://goo.gl/5fpeuG
- Kurath G. Biotechnology and DNA vaccines for aquatic animals. Rev Sci Tech. 2008; 27: 175-196. Ref.: https://goo.gl/x4zCNW
- Kurath G, Garver KA, Corbeil S, Elliott DG, Anderson ED, et al. Protective immunity and lack of histopathological damage two years after DNA vaccination against infectious hematopoietic necrosis virus in trout. Vaccine. 2006; 24: 345-354. Ref.: https://goo.gl/hZcv4a
- Jorgensen HB, Sorensen P, Cooper GA, Lorenzen E, Lorenzen N, et al. General and family-specific gene expression responses to viral hemorrhagic septicaemia virus infection in rainbow trout (Goniorhynchids’ mykiss). Mol Immunol. 2011; 48: 1046-1058. Ref.: https://goo.gl/uVXYvN
- Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 2008; 25: 341-350. Ref.: https://goo.gl/D9abQS
- Novoa B, Romero A, Mulero V, Rodriguez I, Fernandez I, et al. Zebrafish (Danio rerio) as a model for the study of vaccination against viral haemorrhagic septicemia virus (VHSV). Vaccine. 2006; 24: 5806-5816. Ref.: https://goo.gl/yaa1T9
- Encinas P, Rodriguez-Milla MA, Novoa B, Estepa A, Figueras A, et al. Zebrafish fin immune responses during high mortality infections with viral haemorrhagic septicemia rhabdovirus. A proteomic and transcriptomic approach. BMC Genomics. 2010; 27; 518-534. Ref.: https://goo.gl/s2oAmZ
- Fernandez-Alonso M, Rocha A, Coll JM. DNA vaccination by immersion and ultrasound to trout viral haemorrhagic septicaemia virus. Vaccine. 2001; 30: 3067-3075. Ref.: https://goo.gl/V29nkC
- LaPatra SE, Barone L, Jones GR, Zon LI. Effects on infectious hematopoietic necrosis virus and infectious necrosis virus infection on hematopoietic precursosrs of the zebrafish. Blood Cells Mol Dis. 2000; 26: 445-452. Ref.: https://goo.gl/deXwgi
- Phelan PE, Pressley ME, Witten PE, Mellon MT, Blake S, et al. Characterization of snakehead rhabdovirus infection in zebrafish (Danio rerio). J Virol. 2005; 79: 1842-1852. Ref.: https://goo.gl/nmYVrp
- Sanders GE, Batts WN, Winton JR. Susceptibility of zebrafish (Danio rerio) to a model pathogen, spring viremia of carp virus. Comp Med. 2003; 53: 514-521. Ref.: https://goo.gl/SK5H62
- Harmache A, Leberre M, Droineau S, Giovannini M, Bremont M. Bioluminescence Imaging of Live Infected Salmonids Reveals that the Fin Bases Are the Major Portal of Entry for Novirhabdovirus. J Virol. 2006; 80: 3655-3659. Ref.: https://goo.gl/BVrgSV
- Acosta F, Collet B, Lorenzen N, Ellis AE. Expression of the glycoprotein of viral haemorrhagic septicaemia virus (VHSV) on the surface of the fish cell line RTG-P1 induces type 1 interferon expression in neighboring cells. Fish Shellfish Immunol. 2006; 21: 272-278. Ref.: https://goo.gl/yughpQ
- Tafalla C, Coll J, Secombes CJ. Expression of genes related to the early immune response in rainbow trout (Oncorhynchus mykiss) after viral haemorrhagic septicemia virus (VHSV) infection. Developmental Comparative Immunol. 2005; 29: 615-626. Ref.: https://goo.gl/cqpSaU
- Tafalla C, Chico V, Perez L, Coll JM, Estepa A. In vitro and in vivo differential expression of rainbow trout (Oncorhynchus mykiss) Mx isoforms in response to viral haemorrhagic septicaemia virus (VHSV) G gene, poly I:C and VHSV. Fish Shellfish Immunol. 2007; 23: 210-221. Ref.: https://goo.gl/vvxHSF
- Zhang Z, Swain T, Bogwald J, Dalmo RA, Kumari J. Bath immunostimulation of rainbow trout (Oncorhynchus mykiss) fry induces enhancement of inflammatory cytokine transcripts, while repeated bath induce no changes. Fish Shellfish Immunol. 2009; 26: 677-684. Ref.: https://goo.gl/mJxsE5
- Rajcani J, Mosko T, Rezuchova I. Current developments in viral DNA vaccines: shall they solve the unsolved? Rev Med Virol. 2005; 15: 303-325. Ref.: https://goo.gl/QvpUAJ
- Secombes C. Will advances in fish immunology change vaccination strategies? Fish Shellfish Immunol. 2008; 25: 409-416. Ref.: https://goo.gl/ZjSL4h
- Byon JY, Ohira T, Hirono I, Aoki T. Use of a cDNA microarray to study immunity against viral hemorrhagic septicemia (VHS) in Japanese flounder (Paralichthys olivaceous) following DNA vaccination. Fish Shellfish Immunol. 2005; 18: 135-147. Ref.: https://goo.gl/L2kbwi
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005; 23: 307-336. Ref.: https://goo.gl/YEmjaU
- Salem M, Kenney PB, Rexroad CE III, Yao J. Development of a 37-k high-density oligonucleotide microarray: a new tool for functional genome research in rainbow trout. J Fish Biol. 2008; 72: 2187-2206. Ref.: https://goo.gl/zc85HJ
- Byon JYT, Ohira T, Hirono I, Aoki T. Comparative immune responses in Japanese flounder, Paralichthys olivaceous after vaccination with viral hemorrhagic septicemia virus (VHSV) recombinant glycoprotein and DNA vaccine using a microarray analysis. Vaccine. 2006; 24: 921-930. Ref.: https://goo.gl/5MwbVr
- Williams DR, Li W, Hughes MA, Gonzalez SF, Vernon C, et al. Genomic resources and microarrays for the common carp Cyprinus carpio L. J Fish Biol. 2008; 72: 2095-2117. Ref.: https://goo.gl/XnmZSf
- Salem M, Kenney PB, Rexroad CE, Yao J. Development of a 37 k high-density oligonucleotide microarray: a new tool for functional genome research in rainbow trout. J Fish Biol. 2008; 72: 2187-2206. Ref.: https://goo.gl/xULmr2
- Baron D, Montfort J, Houlgatte R, Fostier A, Guiguen Y. Androgen-induced masculinization in rainbow trout results in a marked dysregulation of early gonadal gene expression profiles. BMC Genomics. 2007; 8: 357. Ref.: https://goo.gl/XNnC6r
- Mazurais D, Montfort J, Delalande C, Le Gac F. Transcriptional analysis of testis maturation using trout cDNA microarrays. Gen Comp Endocr. 2005; 142: 143-152. Ref.: https://goo.gl/xVnVjM
- Knoll-Gellida A, André M, Gattegno T, Jean Forgue, Arie Admon, et al. Molecular phenotype of zebrafish ovarian follicle by serial analysis of gene expression and proteomic profiling, and comparison with the transcriptomes of other animals. BMC Genomics. 2006; 7: 46. Ref.: https://goo.gl/aN9zcW
- Juanchich Amélie, Aurélie Le Cam, Jérôme Montfort, Yann Guiguen, Julien Bobe. Identification of Differentially Expressed miRNAs and Their Potential Targets During Fish Ovarian Development. Biol Reprod. 2013; 88: 1-11. Ref.: https://goo.gl/7k3wPy
- Bobe J, Montfort J, Nguyen T, Fostier A. Identification of new participants in the rainbow trout (Oncorhynchus mykiss) oocyte maturation and ovulation processes using cDNA microarrays. Reprod Biol Endocrinol. 2006; 4: 39-10. Ref.: https://goo.gl/joqcWo
- Knoll-Gellida A, Andre M, Gattegno T, Forgue J, Admon A, et al. Molecular phenotype of zebrafish ovarian follicle by serial analysis of gene expression and proteomic profiling, and comparison with the transcriptomes of other animals. BMC Genomics. 2006; 7: 46-10. Ref.: https://goo.gl/TTXbzX
- Von Schalburg KR, Rise ML, Brown GD, Davidson WS, Koop BF. A comprehensive survey of the genes involved in maturation and development of the rainbow trout ovary. Biol Reprod. 2005, 72: 687-699. Ref.: https://goo.gl/tBwFCY
- Bertucci F, Bernard K, Loriod B, Chang YC, Granjeaud S, et al. Sensitivity issues in DNA array-based expression measurements and performance of nylon microarrays for small samples. Hum Mol Genet. 1999; 8: 1715-1722. Ref: https://goo.gl/R5pNak
- Bonnet E, Jalabert B, Bobe J. A 3-day in vitro stage of rainbow trout (Oncorhynchus mykiss) unfertilized eggs in coelomic fluid at 12°C does not affect developmental success. Cybium. 2003; 27: 47-51. Ref.: https://goo.gl/nzr4yi
- Rime H, Guitton N, Pineau C, Bonnet E, Bobe J, et al. 2004. Post-ovulatory ageing and egg quality: A proteomic analysis of rainbow trout coelomic fluid. Endocrinol. 2004; 2: 26. Ref.: https://goo.gl/7CLHpJ
- Salem M, Kenney PB, Rexroad CE, Yao J. Development of a 37 k high-density oligonucleotide microarray: a new tool for functional genome research in rainbow trout. J Fish Biol. 2008; 72: 2187-2206. Ref.: https://goo.gl/qDcKYh